Abstract
Background
In recent years, long noncoding RNAs (LncRNAs) have been found to play an important role in type 2 diabetes mellitus. However, research on the relationship between LncRNAs and prediabetes is still emerging.
Objectives
The study aim was to screen differently expressed LncRNAs and understand their localization and function in patients with prediabetes.
Methods
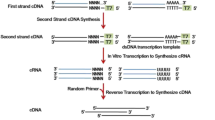
We used microarray analysis to screen LncRNAs in prediabetes participants.To further clarify the localization and function of the expressed mRNAs, we used gene ontology analysis and pathway analysis. Then, internal validations were performed using individual quantitative real-time polymerase chain reaction (qRT-PCR) assays.
Results
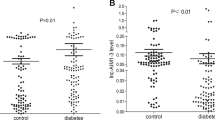
We identified 55 differently expressed LncRNAs and 36 mRNAs in prediabetes participants comparing with controls. Gene ontology analysis indicated that the most enriched transcript terms were multicellular organismal process, plasma membrane, and binding. Pathway analysis indicated that the differently expressed mRNAs were involved in processes such as starch and sucrose metabolism, pantothenate and coenzyme A biosynthesis, and nicotinate and nicotinamide metabolism. The qRT-PCR results showed a trend consistent with the microarray results in 30 patients and 30 healthy controls.
Conclusions
We found aberrantly expressed LncRNAs and mRNAs in prediabetes subjects, and demonstrated that these LncRNAs are involved in the entire prediabetes biological process.





Similar content being viewed by others
References
P. Kapranov, J. Cheng, S. Dike et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316(5830), 1484–1488 (2007)
A. Fatica, I. Bozzoni, Long non-coding RNAs: new players in cell differentiation and development. Nat. Rev. Genet. 15(1), 7–21 (2013)
M. Cesana, D. Cacchiarelli, I. Legnini et al. A long noncoding RNA controls muscle differentiation by functioning as a competing endogenous RNA. Cell 147(2), 358–369 (2011)
C.P. Chang, P. Han, Epigenetic and LncRNAs regulation of cardiac pathophysiology. Biochimica et Biophysica Acta (BBA) Mol. Cell Res. 1863, S0167488916300490 (2016)
T. Nagano, P. Fraser, No-nonsense functions for long noncoding RNAs. Cell 145(2), 178–181 (2011)
R.A. Gupta, N. Shah, K.C. Wang et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464(7291), 1071–1076 (2010)
F. Leti, J.K. Distefano, Long noncoding RNAs as diagnostic and therapeutic targets in type 2 diabetes and related complications. Genes 8(8), 207 (2017)
T. Arita, D. Ichikawa, H. Konishi et al. Circulating long non-coding RNAs in plasma of patients with gastric cancer. Anticancer Res. 33(8), 3185–3193 (2013)
R.A. Boon, N. Jaé, L. Holdt et al. Long noncoding RNAs: from clinical genetics to therapeutic targets? J. Am. Coll. Cardiol. 67(10), 1214–1226 (2016)
D. De Gonzalo-Calvo, F. Kenneweg, C. Bang et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci. Rep. 6, 37354 (2016)
L. Wang, P. Gao, M. Zhang et al. Prevalence and ethnic pattern of diabetes and prediabetes in China in 2013. JAMA 317(24), 2515 (2017)
J. Lindstrom, A. Louheranta, M. Mannelin et al. The Finnish Diabetes Prevention Study (DPS): lifestyle intervention and 3-year results on diet and physical activity. Diabetes Care 26(12), 3230–3236 (2003)
X. Li, Z. Zhao, C. Gao et al. The diagnostic value of whole blood lncRNAs ENST00000550337.1 for pre-diabetes and type 2 diabetes mellitus. Exp. Clin. Endocrinol. Diabetes 125, (2017). https://doi.org/10.1055/s-0043-100018
Chinese Diabetes Society, Guidelines for the prevention and control of type 2 diabetes in China (2017). Chin. J. Diabetes Mellit. 10(1), 4–67 (2018)
M. Ignasi, I. Akerman, M.V.D. Bunt et al. Human β cell transcriptome analysis uncovers LncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab. 16(4), 435–448 (2012)
A.A. Thomas, B. Feng, S. Chakrabarti, ANRIL: a regulator of VEGF in diabetic retinopathy. Invest. Ophthalmol. Vis. Sci. 58(1), 470–80 (2017)
Y. Kotake, T. Nakagawa, K. Kitagawa et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15p15INK4B tumor suppressor gene[J]. Oncogene 30, 1956–1962 (2011)
Y.H. Zhao, T.F. Ji, Q. Luo et al. Long non-coding RNA H19 induces hippocampal neuronal apoptosis via Wnt signaling in a streptozotocin-induced rat model of diabetes mellitus. Oncotarget 8(39), 64827–64839 (2017)
Y.S. Kanwar, X. Pan, S. Lin et al. Imprinted mesodermal specific transcript (MEST) and H19 genes in renal development and diabetes. Kidney Int. 63(5), 1658–1670 (2003)
L.H. You, N. Wang, D.D. Yin et al. Downregulation of long noncoding RNA Meg3 affects insulin synthesis and secretion in mouse pancreatic beta cells. J. Cell. Physiol. 231(4), 852–862 (2016)
G. Carter, B. Miladinovic, A.A. Patel et al. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin. 4, 102–107 (2015)
A.M. Lucrecia, K. Mahdieh, E. Elena et al. Role of MicroRNA 1207-5P and its host gene, the long non-coding RNA Pvt1, as mediators of extracellular matrix accumulation in the kidney: implications for diabetic nephropathy. PLoS ONE 8(10), e77468 (2013)
Y. Ruan, N. Lin, Q. Ma et al. Circulating LncRNAs analysis in patients with type 2 diabetes reveals novel genes influencing glucose metabolism and islet β-cell function. Cell. Physiol. Biochem. 46, 335–350 (2018)
X. Wang, X. Chang, P. Zhang et al. Aberrant expression of long non-coding RNAs in newly diagnosed type 2 diabetes indicates potential roles in chronic inflammation and insulin resistance. Cell. Physiol. Biochem. 43(6), 2367–2378 (2017)
Y.S. Mao, H. Sunwoo, B. Zhang et al. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol. 13(1), 95 (2011)
G. Biamonti, C. Vourc'h, Nuclear stress bodies. Cold Spring Harbor Perspect. Biol. 2(6), a000695 (2010)
J.M. Engreitz, N. Ollikainen, M. Guttman, Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat. Rev. Mol. Cell Biol. 17, 756–770 (2016)
H.M. Krause, New and prospective roles for LncRNAs in organelle formation and function. Trends Genet. 34, S0168952518301124 (2018)
A. Panda, I. Grammatikakis, J.H. Yoon et al. Posttranscriptional regulation of insulin family ligands and receptors. Int. J. Mol. Sci. 14(9), 19202–19229 (2013)
A. Keniry, D. Oxley, P. Monnier et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 14(7), 659–665 (2012)
F. Santoro, D. Mayer, R.M. Klement et al. Imprinted Igf2r silencing depends on continuous Airn LncRNAs expression and is not restricted to a developmental window. Development 140(6), 1184–1195 (2013)
M. Navab, N. Gharavi, A.D. Watson, Inflammation and metabolic disorders. Curr. Opin. Clin. Nutr. Metab. Care 11(4), 459–464 (2008)
S.R. Zatalia, H. Sanusi, The role of antioxidants in the pathophysiology, complications, and management of diabetes mellitus. Acta Med. Indonesiana 45(2), 141–147 (2013)
A.R. Aroor, S. Mckarns, V.G. Demarco et al. Maladaptive immune and inflammatory pathways lead to cardiovascular insulin resistance. Metabolism 62(11), 1543–1552 (2013)
Acknowledgements
We thank Diane Williams, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This work was supported by a grant from the Preventive Medicine research projects of Jiangsu Province Health Department in 2018 (Y2018016) and the Medical Youth Talent of the Project of Invigorating Health Care through Science in Jiangsu Province (QNRC2016375), the Preventive Medicine Research Projects of Jiangsu Province Health Department in 2015 (Y2015010), and the Science and Technology projects of Xuzhou City in 2015 (KC15SM046). The researchers had no relationships with the funder. The study funding had no influence on the study design, data collection, analysis, interpretation of data, writing of the report, or decision to submit the article for publication.
Author information
Authors and Affiliations
Contributions
PZ designed the study, acquired and analyzed data, and drafted and reviewed the manuscript. XZ and YD conducted experiments, reviewed the manuscript, and contributed to the method. ZD conceived and investigated the study, analyzed data, and reviewed the manuscript. CQ and TL researched data, contributed to the discussion, and reviewed the manuscript. PC analyzed data and contributed to the introduction. PL conceived and designed the study, acquired and analyzed data, drafted and reviewed the manuscript, obtained funding, provided administrative support, and supervised the study. PL is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis, reviewed and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was conducted with approval from the ethics committee of Xuzhou Center for Disease Control and Prevention. All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, P., Zhu, X., Du, Y. et al. Screening and functional studies of long noncoding RNA in subjects with prediabetes. Endocrine 68, 296–305 (2020). https://doi.org/10.1007/s12020-020-02226-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-020-02226-3