Abstract
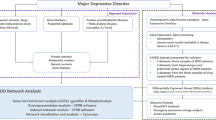
Major depressive disorder is a common debilitating disorder that is associated with increased morbidity and mortality. However, the molecular mechanism underlying depression remains largely unknown. The current study investigated the association of depression with blood gene expression using data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Depression was measured by the geriatric depression scale, and the blood gene expression was measured by the Affymetrix Human Genome U219 Array. Linear regression was used to test the association between gene expression and depression, and the model was adjusted for age and sex. A total of 671 participants were included in our study (mean age 75 ± 8 years, 43.2% women). We found three genes were associated with depression, including COL1A2 (P = 8.9 × 10−8), RNF150 (P = 1.4 × 10−7) and CTGF (P = 8.3 × 10−7). An interaction network was built, and the pathway analysis indicated that many depression-related genes were involved in the neurotrophin signaling pathway (P = 2.1 × 10−7). Future studies are necessary to validate our findings and further investigate potential mechanism of depression.


Similar content being viewed by others
References
Berglund, M., & Nilsson, K. (1987). Mortality in severe depression. A prospective study including 103 suicides. Acta Psychiatrica Scandinavica, 76(4), 372–380.
Boomsma, D. I., Willemsen, G., Sullivan, P. F., Heutink, P., Meijer, P., Sondervan, D., et al. (2008). Genome-wide association of major depression: description of samples for the GAIN Major Depressive Disorder Study: NTR and NESDA biobank projects. European Journal of Human Genetics, 16(3), 335–342. https://doi.org/10.1038/sj.ejhg.5201979.
Broadhead, W. E., Blazer, D. G., George, L. K., & Tse, C. K. (1990). Depression, disability days, and days lost from work in a prospective epidemiologic survey. JAMA, 264(19), 2524–2528.
Bryant, A. J., Carrick, R. P., McConaha, M. E., Jones, B. R., Shay, S. D., Moore, C. S., et al. (2016). Endothelial HIF signaling regulates pulmonary fibrosis-associated pulmonary hypertension. American Journal of Physiology Lung Cellular and Molecular Physiology, 310(3), L249–L262. https://doi.org/10.1152/ajplung.00258.2015.
Cavanagh, J. T., Carson, A. J., Sharpe, M., & Lawrie, S. M. (2003). Psychological autopsy studies of suicide: a systematic review. Psychological Medicine, 33(3), 395–405.
Chu, C. Y., Chang, C. C., Prakash, E., & Kuo, M. L. (2008). Connective tissue growth factor (CTGF) and cancer progression. Journal of Biomedicine Science, 15(6), 675–685. https://doi.org/10.1007/s11373-008-9264-9.
Cowley, M. J., Pinese, M., Kassahn, K. S., Waddell, N., Pearson, J. V., Grimmond, S. M., et al. (2012). PINA v2.0: mining interactome modules. [Research Support, Non-U.S. Gov’t]. Nucleic Acids Research. 40(Database issue), D862–D865. https://doi.org/10.1093/nar/gkr967.
Craddock, N., & Forty, L. (2006). Genetics of affective (mood) disorders. European Journal of Human Genetics, 14(6), 660–668. https://doi.org/10.1038/sj.ejhg.5201549.
Ding, Y., Niu, H., Yang, H., Sun, P., Chen, Y., Duan, M., et al. (2015). EGLN2 and RNF150 genetic variants are associated with chronic obstructive pulmonary disease risk in the Chinese population. International Journal of Chronic Obstructive Pulmonary Disease, 10, 145–151. https://doi.org/10.2147/COPD.S73031.
Farmer, A., Harris, T., Redman, K., Sadler, S., Mahmood, A., & McGuffin, P. (2000). Cardiff depression study. A sib-pair study of life events and familiality in major depression. The British Journal of Psychiatry, 176, 150–155.
Fonseca, C., Lindahl, G. E., Ponticos, M., Sestini, P., Renzoni, E. A., Holmes, A. M., et al. (2007). A polymorphism in the CTGF promoter region associated with systemic sclerosis. New England Journal of Medicine, 357(12), 1210–1220. https://doi.org/10.1056/NEJMoa067655.
Granel, B., Argiro, L., Hachulla, E., Fajardy, I., Weiller, P. J., Durand, J. M., et al. (2010). Association between a CTGF gene polymorphism and systemic sclerosis in a French population. The Journal of Rheumatology, 37(2), 351–358. https://doi.org/10.3899/jrheum.090290.
Greenberg, P. E., Stiglin, L. E., Finkelstein, S. N., & Berndt, E. R. (1993). The economic burden of depression in 1990. The Journal of Clinical Psychiatry, 54(11), 405–418.
GTEx Consortium. (2015). Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science, 348(6235), 648–660. https://doi.org/10.1126/science.1262110.
Ideker, T., Ozier, O., Schwikowski, B., & Siegel, A. F. (2002). Discovering regulatory and signalling circuits in molecular interaction networks. Bioinformatics, 18(Suppl 1), S233–S240.
Jacobson, A., & Cunningham, J. L. (2012). Connective tissue growth factor in tumor pathogenesis. Fibrogenesis Tissue Repair. 5(Suppl 1), S8. https://doi.org/10.1186/1755-1536-5-S1-S8.
Jia, P., Zheng, S., Long, J., Zheng, W., & Zhao, Z. (2011). dmGWAS: dense module searching for genome-wide association studies in protein-protein interaction networks. Bioinformatics, 27(1), 95–102.
Kendler, K. S., Gatz, M., Gardner, C. O., & Pedersen, N. L. (2005). Age at onset and familial risk for major depression in a Swedish national twin sample. Psychological Medicine, 35(11), 1573–1579. https://doi.org/10.1017/S0033291705005714.
Kendler, K. S., Neale, M. C., Kessler, R. C., Heath, A. C., & Eaves, L. J. (1993). The lifetime history of major depression in women. Reliability of diagnosis and heritability. Archives of General Psychiatry, 50(11), 863–870.
Kessler, R. C., Berglund, P., Demler, O., Jin, R., Merikangas, K. R., & Walters, E. E. (2005). Lifetime prevalence and age-of-onset distributions of DSM-IV disorders in the National Comorbidity Survey Replication. Archives of General Psychiatry, 62(6), 593–602. https://doi.org/10.1001/archpsyc.62.6.593.
Lewis, C. M., Ng, M. Y., Butler, A. W., Cohen-Woods, S., Uher, R., Pirlo, K., et al. (2010). Genome-wide association study of major recurrent depression in the U.K. population. American Journal of Psychiatry, 167(8), 949–957. https://doi.org/10.1176/appi.ajp.2010.09091380.
Lopez, A. D., & Murray, C. C. (1998). The global burden of disease, 1990–2020. Nature Medicine, 4(11), 1241–1243. https://doi.org/10.1038/3218.
Lopez-Leon, S., Janssens, A. C., Gonzalez-Zuloeta Ladd, A. M., Del-Favero, J., Claes, S. J., Oostra, B. A., et al. (2008). Meta-analyses of genetic studies on major depressive disorder. Molecular Psychiatry, 13(8), 772–785. https://doi.org/10.1038/sj.mp.4002088.
Marc, L. G., Raue, P. J., & Bruce, M. L. (2008). Screening performance of the 15-item geriatric depression scale in a diverse elderly home care population. The American Journal of Geriatric Psychiatry, 16(11), 914–921. https://doi.org/10.1097/JGP.0b013e318186bd67.
McGuffin, P., Katz, R., Watkins, S., & Rutherford, J. (1996). A hospital-based twin register of the heritability of DSM-IV unipolar depression. Archives of General Psychiatry, 53(2), 129–136.
Miao, X., Chen, X., Xie, Z., & Lin, H. (2018). Tissue-specific network analysis of genetic variants associated with coronary artery disease. Scientific Reports, 8(1), 11492. https://doi.org/10.1038/s41598-018-29904-7.
Muglia, P., Tozzi, F., Galwey, N. W., Francks, C., Upmanyu, R., Kong, X. Q., et al. (2010). Genome-wide association study of recurrent major depressive disorder in two European case-control cohorts. Molecular Psychiatry, 15(6), 589–601. https://doi.org/10.1038/mp.2008.131.
Peall, K. J., Kurian, M. A., Wardle, M., Waite, A. J., Hedderly, T., Lin, J. P., et al. (2014). SGCE and myoclonus dystonia: motor characteristics, diagnostic criteria and clinical predictors of genotype. Journal of Neurology, 261(12), 2296–2304. https://doi.org/10.1007/s00415-014-7488-3.
Petersen, R. C., Aisen, P. S., Beckett, L. A., Donohue, M. C., Gamst, A. C., Harvey, D. J., et al. (2010). Alzheimer’s Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology, 74(3), 201–209. https://doi.org/10.1212/WNL.0b013e3181cb3e25.
Pope, F. M., Kendall, B. E., Slapak, G. I., Kapoor, R., McDonald, W. I., Compston, D. A., et al. (1991). Type III collagen mutations cause fragile cerebral arteries. British Journal of Neurosurgery, 5(6), 551–574.
Saykin, A. J., Shen, L., Yao, X., Kim, S., Nho, K., Risacher, S. L., et al. (2015). Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dementia, 11(7), 792–814. https://doi.org/10.1016/j.jalz.2015.05.009.
Schule, B., Kock, N., Svetel, M., Dragasevic, N., Hedrich, K., De Carvalho Aguiar, P., et al. (2004). Genetic heterogeneity in ten families with myoclonus-dystonia. Journal of Neurology Neurosurgery and Psychiatry, 75(8), 1181–1185. https://doi.org/10.1136/jnnp.2003.027177.
Shi, J., Potash, J. B., Knowles, J. A., Weissman, M. M., Coryell, W., Scheftner, W. A., et al. (2011). Genome-wide association study of recurrent early-onset major depressive disorder. Molecular Psychiatry, 16(2), 193–201. https://doi.org/10.1038/mp.2009.124.
Shyn, S. I., Shi, J., Kraft, J. B., Potash, J. B., Knowles, J. A., Weissman, M. M., et al. (2011). Novel loci for major depression identified by genome-wide association study of Sequenced Treatment Alternatives to Relieve Depression and meta-analysis of three studies. Molecular Psychiatry, 16(2), 202–215. https://doi.org/10.1038/mp.2009.125.
Sullivan, P. F., de Geus, E. J., Willemsen, G., James, M. R., Smit, J. H., Zandbelt, T., et al. (2009). Genome-wide association for major depressive disorder: a possible role for the presynaptic protein piccolo. Molecular Psychiatry, 14(4), 359–375. https://doi.org/10.1038/mp.2008.125.
Sullivan, P. F., Neale, M. C., & Kendler, K. S. (2000). Genetic epidemiology of major depression: review and meta-analysis. American Journal of Psychiatry, 157(10), 1552–1562. https://doi.org/10.1176/appi.ajp.157.10.1552.
Wang, J., Duncan, D., Shi, Z., & Zhang, B. (2013). WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Research, 41(Web Server issue), W77–W83. https://doi.org/10.1093/nar/gkt439.
Waraich, P., Goldner, E. M., Somers, J. M., & Hsu, L. (2004). Prevalence and incidence studies of mood disorders: a systematic review of the literature. Canadian Journal of Psychiatry, 49(2), 124–138. https://doi.org/10.1177/070674370404900208.
Weissman, M. M., Bland, R., Joyce, P. R., Newman, S., Wells, J. E., & Wittchen, H. U. (1993). Sex differences in rates of depression: cross-national perspectives. Journal of Affective Disorders, 29(2–3), 77–84.
Wells, K. B., Stewart, A., Hays, R. D., Burnam, M. A., Rogers, W., Daniels, M., et al. (1989). The functioning and well-being of depressed patients. Results from the Medical Outcomes Study. JAMA, 262(7), 914–919.
Wilhelm, K., Mitchell, P., Slade, T., Brownhill, S., & Andrews, G. (2003). Prevalence and correlates of DSM-IV major depression in an Australian national survey. Journal of Affective Disorders, 75(2), 155–162.
Wray, N. R., Pergadia, M. L., Blackwood, D. H., Penninx, B. W., Gordon, S. D., Nyholt, D. R., et al. (2012). Genome-wide association study of major depressive disorder: new results, meta-analysis, and lessons learned. Molecular Psychiatry, 17(1), 36–48. https://doi.org/10.1038/mp.2010.109.
Zimprich, A., Grabowski, M., Asmus, F., Naumann, M., Berg, D., Bertram, M., et al. (2001). Mutations in the gene encoding epsilon-sarcoglycan cause myoclonus-dystonia syndrome. Nature Genetics, 29(1), 66–69. https://doi.org/10.1038/ng709.
Acknowledgements
This project was supported by the National Natural Science Foundation of China (Grant No. 81673977 and 81673863), the Shanghai Science and Technology Commission of China (Grant No: 18511107700), and the Shanghai High-level University Development Project. It was also supported by the Boston University Digital Health Initiative, Boston University Alzheimer’s Disease Center Pilot Grant, and the National Center for Advancing Translational Sciences, National Institutes of Health, through BU-CTSI Grant Number 1UL1TR001430. The views expressed in this manuscript are those of the authors and do not necessarily represent the views of the National Institutes of Health or the U.S. Department of Health and Human Services. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (http://www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Author information
Authors and Affiliations
Consortia
Contributions
XM and HL initiated the study and drafted the manuscript. BF, RL, SZ and BZ performed the analyses and critically reviewed the manuscript. All authors approve the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical Approval
The institutional review boards of all sites participating in the ADNI provided review and approval of the ADNI data collection protocol. All participants provided written consent.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.usc.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at http://adni.loni.usc.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Miao, X., Fan, B., Li, R. et al. Network Analysis of Depression-Related Transcriptomic Profiles. Neuromol Med 21, 143–149 (2019). https://doi.org/10.1007/s12017-019-08527-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12017-019-08527-9