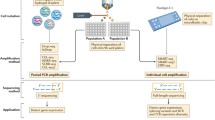
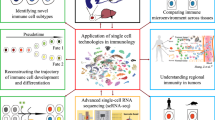
Abstract
Complex composition is one of the most important features of the immune system, involving many types of organs, tissues, cells, and molecules that perform immune functions. The normal function of each component of the immune system is the guarantee for maintaining the relatively stable immune function of the body. When the self-immune tolerance mechanism of the body is unregulated or destroyed, the immune system reacts to autoantigens, resulting in damage to self-tissues and organs or an immunopathological state with abnormal functions. Autoimmune diseases are diverse, and their pathogenesis is complicated. Various immune cells and their interactions play significant roles in the occurrence and development of diseases. The solution to heterogeneity of immune cells is the basic science and translational understanding of how genes and the environment interact to induce disease so that we can develop personalized medicine, a goal that has to this point eluded scientists. Single-cell RNA sequencing (scRNA-Seq) refers to a new technique allowing high-throughput sequencing analysis of the whole transcriptome to reveal the gene expression status of individual cells. It has emerged as an indispensable tool in the field of life science research, and can help identify the complex mechanism of cell heterogeneity, discover new cell subsets, and help uncover the molecular mechanisms of pathogenesis, the evolution of disorders, and drug resistance. This information can provide us with new strategies for diagnosis and prognostic evaluation, as well as monitoring treatment responses. In this review, we summarize the crucial experimental procedures used for single-cell RNA sequencing, and the current applications of this technique to study autoimmune diseases are described in detail. This technique will be widely used in more in-depth studies of autoimmune diseases and will contribute to the diagnosis and therapies of these disorders.

Similar content being viewed by others
References
Coskun AF, Eser U, Islam S (2016) Cellular identity at the single-cell level. Mol BioSyst 12(10):2965–2979
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5):377–382
Islam S, Kjallquist U, Moliner A, Zajac P, Fan JB, Lonnerberg P, Linnarsson S (2011) Characterization of the single-cell transcriptional landscape by highly multiplex RNA-seq. Genome Res 21(7):1160–1167
Hashimshony T, Wagner F, Sher N, Yanai I (2012) CEL-Seq: single-cell RNA-Seq by multiplexed linear amplification. Cell Rep 2(3):666–673
Ramskold D et al (2012) Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol 30(8):777–782
Pan X, Durrett RE, Zhu H, Tanaka Y, Li Y, Zi X, Marjani SL, Euskirchen G, Ma C, LaMotte RH, Park IH, Snyder MP, Mason CE, Weissman SM (2013) Two methods for full-length RNA sequencing for low quantities of cells and single cells. Proc Natl Acad Sci U S A 110(2):594–599
Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, Sandberg R (2014) Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc 9(1):171–181
Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I (2014) Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science 343(6172):776–779
Fan HC, Fu GK, Fodor SP (2015) Expression profiling. Combinatorial labeling of single cells for gene expression cytometry. Science 347(6222):1258367
Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161(5):1202–1214
Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5):1187–1201
Cao J et al (2017) Comprehensive single-cell transcriptional profiling of a multicellular organism. Science (New York, N.Y.) 357(6352):661–667
Gierahn TM, Wadsworth MH II, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK (2017) Seq-well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14(4):395–398
Single-cell profiling of the developing mouse brain and spinal cord with split-pool barcoding. 2018
Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, Huang D, Xu Y, Huang W, Jiang M, Jiang X, Mao J, Chen Y, Lu C, Xie J, Fang Q, Wang Y, Yue R, Li T, Huang H, Orkin SH, Yuan GC, Chen M, Guo G (2018) Mapping the mouse cell atlas by Microwell-Seq. Cell 172(5):1091–1107 e17
Chen H, Ye F, Guo G (2019) Revolutionizing immunology with single-cell RNA sequencing. Cell Mol Immunol 16(3):242–249
Huang XT, Li X, Qin PZ, Zhu Y, Xu SN, Chen JP (2018) Technical advances in single-cell RNA sequencing and applications in normal and malignant hematopoiesis. Front Oncol 8:582
Kivioja T, Vähärautio A, Karlsson K, Bonke M, Enge M, Linnarsson S, Taipale J (2011) Counting absolute numbers of molecules using unique molecular identifiers. Nat Methods 9(1):72–74
Islam S, Zeisel A, Joost S, la Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S (2014) Quantitative single-cell RNA-seq with unique molecular identifiers. Nat Methods 11(2):163–166
Guo MT, Rotem A, Heyman JA, Weitz DA (2012) Droplet microfluidics for high-throughput biological assays. Lab Chip 12(12):2146–2155
Morris J, Singh JM, Eberwine JH (2011) Transcriptome analysis of single cells. Journal of Visualized Experiments : JoVE 50
Guo F, Li L, Li J, Wu X, Hu B, Zhu P, Wen L, Tang F (2017) Single-cell multi-omics sequencing of mouse early embryos and embryonic stem cells. Cell Res 27(8):967–988
Nakamura N et al (2007) Laser capture microdissection for analysis of single cells. Methods in Molecular Medicine 132:11–18
Emmert-Buck MR et al (1996) Laser capture microdissection. Science (New York, N.Y.) 274(5289):5289
Chen J, Suo S, Tam PPL, Han JDJ, Peng G, Jing N (2017) Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nat Protoc 12(3):566–580
Maciorowski Z, Chattopadhyay PK, Jain P (2017) Basic multicolor flow cytometry. Curr Protoc Immunol 117:541–5438
Adan A, Alizada G, Kiraz Y, Baran Y, Nalbant A (2017) Flow cytometry: basic principles and applications. Crit Rev Biotechnol 37(2):163–176
Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, Zabala M, Bueno J, Neff NF, Wang J, Shelton AA, Visser B, Hisamori S, Shimono Y, van de Wetering M, Clevers H, Clarke MF, Quake SR (2011) Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol 29(12):1120–1127
Wu AR, Neff NF, Kalisky T, Dalerba P, Treutlein B, Rothenberg ME, Mburu FM, Mantalas GL, Sim S, Clarke MF, Quake SR (2014) Quantitative assessment of single-cell RNA-sequencing methods. Nat Methods 11(1):41–46
Joensson HN, Andersson Svahn H (2012) Droplet microfluidics--a tool for single-cell analysis. Angewandte Chemie (International ed. in English) 51(49):12176–12192
Valihrach L, Androvic P, Kubista M (2018) Platforms for single-cell collection and analysis. Int J Mol Sci 19(3):807
Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, Ramalingam N, Sun G, Thu M, Norris M, Lebofsky R, Toppani D, Kemp DW II, Wong M, Clerkson B, Jones BN, Wu S, Knutsson L, Alvarado B, Wang J, Weaver LS, May AP, Jones RC, Unger MA, Kriegstein AR, West JAA (2014) Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat Biotechnol 32(10):1053–1058
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, Gregory MT, Shuga J, Montesclaros L, Underwood JG, Masquelier DA, Nishimura SY, Schnall-Levin M, Wyatt PW, Hindson CM, Bharadwaj R, Wong A, Ness KD, Beppu LW, Deeg HJ, McFarland C, Loeb KR, Valente WJ, Ericson NG, Stevens EA, Radich JP, Mikkelsen TS, Hindson BJ, Bielas JH (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun 8:14049
Han X, Zhou Z, Fei L, Sun H, Wang R, Chen Y, Chen H, Wang J, Tang H, Ge W, Zhou Y, Ye F, Jiang M, Wu J, Xiao Y, Jia X, Zhang T, Ma X, Zhang Q, Bai X, Lai S, Yu C, Zhu L, Lin R, Gao Y, Wang M, Wu Y, Zhang J, Zhan R, Zhu S, Hu H, Wang C, Chen M, Huang H, Liang T, Chen J, Wang W, Zhang D, Guo G (2020) Construction of a human cell landscape at single-cell level. Nature 581(7808):303–309
Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH, Carbone L, Steemers FJ, Adey A (2017) Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14(3):302–308
Zhang F et al (2019) Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol 20(7):928–942
Mizoguchi F, Slowikowski K, Wei K, Marshall JL, Rao DA, Chang SK, Nguyen HN, Noss EH, Turner JD, Earp BE, Blazar PE, Wright J, Simmons BP, Donlin LT, Kalliolias GD, Goodman SM, Bykerk VP, Ivashkiv LB, Lederer JA, Hacohen N, Nigrovic PA, Filer A, Buckley CD, Raychaudhuri S, Brenner MB (2018) Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun 9(1):789
Der E et al (2019) Tubular cell and keratinocyte single-cell transcriptomics applied to lupus nephritis reveal type I IFN and fibrosis relevant pathways. Nat Immunol 20(7):915–927
Papalexi E, Satija R (2018) Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol 18(1):35–45
Stephenson W, Donlin LT, Butler A, Rozo C, Bracken B, Rashidfarrokhi A, Goodman SM, Ivashkiv LB, Bykerk VP, Orange DE, Darnell RB, Swerdlow HP, Satija R (2018) Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat Commun 9(1):791
Lu DR, McDavid AN, Kongpachith S, Lingampalli N, Glanville J, Ju CH, Gottardo R, Robinson WH (2018) T cell-dependent affinity maturation and innate immune pathways differentially drive autoreactive B cell responses in rheumatoid arthritis. Arthritis Rheumatol 70(11):1732–1744
Kuo D et al (2019) HBEGF(+) macrophages in rheumatoid arthritis induce fibroblast invasiveness. Sci Transl Med 11(491):eaau8587
Croft AP, Campos J, Jansen K, Turner JD, Marshall J, Attar M, Savary L, Wehmeyer C, Naylor AJ, Kemble S, Begum J, Dürholz K, Perlman H, Barone F, McGettrick HM, Fearon DT, Wei K, Raychaudhuri S, Korsunsky I, Brenner MB, Coles M, Sansom SN, Filer A, Buckley CD (2019) Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570(7760):246–251
Cai S et al (2019) Similar transition processes in synovial fibroblasts from rheumatoid arthritis and osteoarthritis: a single-cell study. J Immunol Res 2019:4080735
Gawel DR, Serra-Musach J, Lilja S, Aagesen J, Arenas A, Asking B, Bengnér M, Björkander J, Biggs S, Ernerudh J, Hjortswang H, Karlsson JE, Köpsen M, Lee EJ, Lentini A, Li X, Magnusson M, Martínez-Enguita D, Matussek A, Nestor CE, Schäfer S, Seifert O, Sonmez C, Stjernman H, Tjärnberg A, Wu S, Åkesson K, Shalek AK, Stenmarker M, Zhang H, Gustafsson M, Benson M (2019) A validated single-cell-based strategy to identify diagnostic and therapeutic targets in complex diseases. Genome Med 11(1):47
Culemann S, Grüneboom A, Nicolás-Ávila JÁ, Weidner D, Lämmle KF, Rothe T, Quintana JA, Kirchner P, Krljanac B, Eberhardt M, Ferrazzi F, Kretzschmar E, Schicht M, Fischer K, Gelse K, Faas M, Pfeifle R, Ackermann JA, Pachowsky M, Renner N, Simon D, Haseloff RF, Ekici AB, Bäuerle T, Blasig IE, Vera J, Voehringer D, Kleyer A, Paulsen F, Schett G, Hidalgo A, Krönke G (2019) Locally renewing resident synovial macrophages provide a protective barrier for the joint. Nature 572(7771):670–675
Hasegawa T, Kikuta J, Sudo T, Matsuura Y, Matsui T, Simmons S, Ebina K, Hirao M, Okuzaki D, Yoshida Y, Hirao A, Kalinichenko VV, Yamaoka K, Takeuchi T, Ishii M (2019) Identification of a novel arthritis-associated osteoclast precursor macrophage regulated by FoxM1. Nat Immunol 20(12):1631–1643
Wei K et al (2020) Notch signalling drives synovial fibroblast identity and arthritis pathology. Nature 582(7811):259–264
Marques S, Zeisel A, Codeluppi S, van Bruggen D, Mendanha Falcao A, Xiao L, Li H, Haring M, Hochgerner H, Romanov RA, Gyllborg D, Munoz-Manchado AB, la Manno G, Lonnerberg P, Floriddia EM, Rezayee F, Ernfors P, Arenas E, Hjerling-Leffler J, Harkany T, Richardson WD, Linnarsson S, Castelo-Branco G (2016) Oligodendrocyte heterogeneity in the mouse juvenile and adult central nervous system. Science 352(6291):1326–1329
Falcao AM et al (2018) Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med 24(12):1837–1844
Hammond TR, Dufort C, Dissing-Olesen L, Giera S, Young A, Wysoker A, Walker AJ, Gergits F, Segel M, Nemesh J, Marsh SE, Saunders A, Macosko E, Ginhoux F, Chen J, Franklin RJM, Piao X, McCarroll SA, Stevens B (2019) Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50(1):253–271 e6
Jordao MJC et al (2019) Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363(6425):eaat7554
Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M (2019) Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature 566(7744):388–392
Beltran E et al (2019) Early adaptive immune activation detected in monozygotic twins with prodromal multiple sclerosis. J Clin Invest 129(11):4758–4768
Schafflick D, Xu CA, Hartlehnert M, Cole M, Schulte-Mecklenbeck A, Lautwein T, Wolbert J, Heming M, Meuth SG, Kuhlmann T, Gross CC, Wiendl H, Yosef N, Meyer zu Horste G (2020) Integrated single cell analysis of blood and cerebrospinal fluid leukocytes in multiple sclerosis. Nat Commun 11(1):247
Wheeler MA, Clark IC, Tjon EC, Li Z, Zandee SEJ, Couturier CP, Watson BR, Scalisi G, Alkwai S, Rothhammer V, Rotem A, Heyman JA, Thaploo S, Sanmarco LM, Ragoussis J, Weitz DA, Petrecca K, Moffitt JR, Becher B, Antel JP, Prat A, Quintana FJ (2020) MAFG-driven astrocytes promote CNS inflammation. Nature 578(7796):593–599
Marisca R, Hoche T, Agirre E, Hoodless LJ, Barkey W, Auer F, Castelo-Branco G, Czopka T (2020) Functionally distinct subgroups of oligodendrocyte precursor cells integrate neural activity and execute myelin formation. Nat Neurosci 23(3):363–374
Esaulova E et al (2020) Single-cell RNA-seq analysis of human CSF microglia and myeloid cells in neuroinflammation. Neurol Neuroimmunol Neuroinflamm 7(4)
Ramesh A, Schubert RD, Greenfield AL, Dandekar R, Loudermilk R, Sabatino JJ Jr, Koelzer MT, Tran EB, Koshal K, Kim K, Pröbstel AK, Banerji D, University of California, San Francisco MS-EPIC Team, Guo CY, Green AJ, Bove RM, DeRisi JL, Gelfand JM, Cree BAC, Zamvil SS, Baranzini SE, Hauser SL, Wilson MR (2020) A pathogenic and clonally expanded B cell transcriptome in active multiple sclerosis. Proc Natl Acad Sci U S A 117(37):22932–22943
Der E et al (2017) Single cell RNA sequencing to dissect the molecular heterogeneity in lupus nephritis. JCI Insight 2(9):e93009
Arazi A et al (2019) The immune cell landscape in kidneys of patients with lupus nephritis. Nat Immunol 20(7):902–914
Lee PY et al (2019) High-dimensional analysis reveals a pathogenic role of inflammatory monocytes in experimental diffuse alveolar hemorrhage. JCI Insight 4(15):e129703
Mistry P, Nakabo S, O’Neil L, Goel RR, Jiang K, Carmona-Rivera C, Gupta S, Chan DW, Carlucci PM, Wang X, Naz F, Manna Z, Dey A, Mehta NN, Hasni S, Dell’Orso S, Gutierrez-Cruz G, Sun HW, Kaplan MJ (2019) Transcriptomic, epigenetic, and functional analyses implicate neutrophil diversity in the pathogenesis of systemic lupus erythematosus. Proc Natl Acad Sci U S A 116(50):25222–25228
Goel RR, Wang X, O’Neil LJ, Nakabo S, Hasneen K, Gupta S, Wigerblad G, Blanco LP, Kopp JB, Morasso MI, Kotenko SV, Yu ZX, Carmona-Rivera C, Kaplan MJ (2020) Interferon lambda promotes immune dysregulation and tissue inflammation in TLR7-induced lupus. Proc Natl Acad Sci U S A 117(10):5409–5419
Nehar-Belaid D, Hong S, Marches R, Chen G, Bolisetty M, Baisch J, Walters L, Punaro M, Rossi RJ, Chung CH, Huynh RP, Singh P, Flynn WF, Tabanor-Gayle JA, Kuchipudi N, Mejias A, Collet MA, Lucido AL, Palucka K, Robson P, Lakshminarayanan S, Ramilo O, Wright T, Pascual V, Banchereau JF (2020) Mapping systemic lupus erythematosus heterogeneity at the single-cell level. Nat Immunol 21(9):1094–1106
Cerosaletti K, Barahmand-pour-Whitman F, Yang J, DeBerg HA, Dufort MJ, Murray SA, Israelsson E, Speake C, Gersuk VH, Eddy JA, Reijonen H, Greenbaum CJ, Kwok WW, Wambre E, Prlic M, Gottardo R, Nepom GT, Linsley PS (2017) Single-cell RNA sequencing reveals expanded clones of islet antigen-reactive CD4(+) T cells in peripheral blood of subjects with type 1 diabetes. J Immunol 199(1):323–335
Sharon N, Vanderhooft J, Straubhaar J, Mueller J, Chawla R, Zhou Q, Engquist EN, Trapnell C, Gifford DK, Melton DA (2019) Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep 27(8):2281–2291 e5
Kallionpaa H et al (2019) Early detection of peripheral blood cell signature in children developing beta-cell autoimmunity at a young age. Diabetes 68(10):2024–2034
Wang D, Wang J, Bai L, Pan H, Feng H, Clevers H, Zeng YA (2020) Long-term expansion of pancreatic islet organoids from resident Procr(+) progenitors. Cell 180(6):1198–1211 e19
Zakharov PN et al (2020) Single-cell RNA sequencing of murine islets shows high cellular complexity at all stages of autoimmune diabetes. J Exp Med 217(6):e20192362
Firestein GS, McInnes IB (2017) Immunopathogenesis of rheumatoid arthritis. Immunity 46(2):183–196
(2018) Rheumatoid arthritis. Nat Rev Dis Primers 4:18002
Smith MD (2011) The normal synovium. Open Rheumatol J 5:100–106
Robinson WH, Lepus CM, Wang Q, Raghu H, Mao R, Lindstrom TM, Sokolove J (2016) Low-grade inflammation as a key mediator of the pathogenesis of osteoarthritis. Nat Rev Rheumatol 12(10):580–592
Smolen JS, Aletaha D, McInnes IB (2016) Rheumatoid arthritis. Lancet 388(10055):2023–2038
Lefevre S, Meier FM, Neumann E, Muller-Ladner U (2015) Role of synovial fibroblasts in rheumatoid arthritis. Curr Pharm Des 21(2):130–141
Huber LC, Distler O, Tarner I, Gay RE, Gay S, Pap T (2006) Synovial fibroblasts: key players in rheumatoid arthritis. Rheumatology (Oxford) 45(6):669–675
Frank-Bertoncelj M, Gay S (2017) Rheumatoid arthritis: TAK-ing the road to suppress inflammation in synovial fibroblasts. Nat Rev Rheumatol 13(3):133–134
Onuora S (2020) Synovial fibroblast expansion in RA is driven by notch signalling. Nat Rev Rheumatol 16(7):349
Udalova IA, Mantovani A, Feldmann M (2016) Macrophage heterogeneity in the context of rheumatoid arthritis. Nat Rev Rheumatol 12(8):472–485
Wu H, Deng Y, Feng Y, Long D, Ma K, Wang X, Zhao M, Lu L, Lu Q (2018) Epigenetic regulation in B-cell maturation and its dysregulation in autoimmunity. Cell Mol Immunol 15(7):676–684
Donlin LT et al (2018) Methods for high-dimensional analysis of cells dissociated from cryopreserved synovial tissue. Arthritis Res Ther 20(1):139
Fonseka CY et al (2018) Mixed-effects association of single cells identifies an expanded effector CD4(+) T cell subset in rheumatoid arthritis. Sci Transl Med 10(463):eaaq0305
Kaul A, Gordon C, Crow MK, Touma Z, Urowitz MB, van Vollenhoven R, Ruiz-Irastorza G, Hughes G (2016) Systemic lupus erythematosus. Nat Rev Dis Primers 2:16039
Vanarsa K, Soomro S, Zhang T, Strachan B, Pedroza C, Nidhi M, Cicalese P, Gidley C, Dasari S, Mohan S, Thai N, Truong VTT, Jordan N, Saxena R, Putterman C, Petri M, Mohan C (2020) Quantitative planar array screen of 1000 proteins uncovers novel urinary protein biomarkers of lupus nephritis. Ann Rheum Dis 79(10):1349–1361
Zhang T, Li H, Vanarsa K, Gidley G, Mok CC, Petri M, Saxena R, Mohan C (2020) Association of urine sCD163 with proliferative lupus nephritis, fibrinoid necrosis, cellular crescents and intrarenal M2 macrophages. Front Immunol 11:671
Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, Gate RE, Mostafavi S, Marson A, Zaitlen N, Criswell LA, Ye CJ (2018) Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36(1):89–94
Dura B et al (2019) scFTD-seq: freeze-thaw lysis based, portable approach toward highly distributed single-cell 3' mRNA profiling. Nucleic Acids Res 47(3):e16
Carmona-Rivera C, Kaplan MJ (2013) Low-density granulocytes: a distinct class of neutrophils in systemic autoimmunity. Semin Immunopathol 35(4):455–463
Rahman S, Sagar D, Hanna RN, Lightfoot YL, Mistry P, Smith CK, Manna Z, Hasni S, Siegel RM, Sanjuan MA, Kolbeck R, Kaplan MJ, Casey KA (2019) Low-density granulocytes activate T cells and demonstrate a non-suppressive role in systemic lupus erythematosus. Ann Rheum Dis 78(7):957–966
Tay SH, Celhar T, Fairhurst AM (2020) Low-density neutrophils in systemic lupus erythematosus. Arthritis Rheum 29:1334
Martinez-Martinez MU, Oostdam DAH, Abud-Mendoza C (2017) Diffuse alveolar hemorrhage in autoimmune diseases. Curr Rheumatol Rep 19(5):27
Martinez-Martinez MU, Abud-Mendoza C (2014) Diffuse alveolar hemorrhage in patients with systemic lupus erythematosus. Clinical manifestations, treatment, and prognosis. Reumatol Clin 10(4):248–253
Filippi M, Bar-Or A, Piehl F, Preziosa P, Solari A, Vukusic S, Rocca MA (2018) Multiple sclerosis. Nat Rev Dis Primers 4(1):43
Schirmer L, Velmeshev D, Holmqvist S, Kaufmann M, Werneburg S, Jung D, Vistnes S, Stockley JH, Young A, Steindel M, Tung B, Goyal N, Bhaduri A, Mayer S, Engler JB, Bayraktar OA, Franklin RJM, Haeussler M, Reynolds R, Schafer DP, Friese MA, Shiow LR, Kriegstein AR, Rowitch DH (2019) Neuronal vulnerability and multilineage diversity in multiple sclerosis. Nature 573(7772):75–82
Falcão AM, van Bruggen D, Marques S, Meijer M, Jäkel S, Agirre E, Samudyata, Floriddia EM, Vanichkina DP, ffrench-Constant C, Williams A, Guerreiro-Cacais AO, Castelo-Branco G (2018) Disease-specific oligodendrocyte lineage cells arise in multiple sclerosis. Nat Med 24(12):1837–1844
Hammond TR et al (2019) Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50(1):253
Li Q, Barres BA (2018) Microglia and macrophages in brain homeostasis and disease. Nat Rev Immunol 18(4):225–242
Katsarou A, Gudbjörnsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, Jacobsen LM, Schatz DA, Lernmark Å (2017) Type 1 diabetes mellitus. Nat Rev Dis Primers 3:17016
Pagliuca FW, Millman JR, Gürtler M, Segel M, van Dervort A, Ryu JH, Peterson QP, Greiner D, Melton DA (2014) Generation of functional human pancreatic β cells in vitro. Cell 159(2):428–439
Veres A, Faust AL, Bushnell HL, Engquist EN, Kenty JHR, Harb G, Poh YC, Sintov E, Gürtler M, Pagliuca FW, Peterson QP, Melton DA (2019) Charting cellular identity during human in vitro β-cell differentiation. Nature 569(7756):368–373
Wang D et al (2020) Long-term expansion of pancreatic islet organoids from resident Procr progenitors. Cell 180(6):1198
Chakravarthy H, Gu X, Enge M, Dai X, Wang Y, Damond N, Downie C, Liu K, Wang J, Xing Y, Chera S, Thorel F, Quake S, Oberholzer J, MacDonald PE, Herrera PL, Kim SK (2017) Converting adult pancreatic islet alpha cells into beta cells by targeting both Dnmt1 and Arx. Cell Metab 25(3):622–634
Chambers DC, Carew AM, Lukowski SW, Powell JE (2019) Transcriptomics and single-cell RNA-sequencing. Respirology 24(1):29–36
Baumer C et al (2018) Exploring DNA quality of single cells for genome analysis with simultaneous whole-genome amplification. Sci Rep 8(1):7476
Macaulay IC, Haerty W, Kumar P, Li YI, Hu TX, Teng MJ, Goolam M, Saurat N, Coupland P, Shirley LM, Smith M, van der Aa N, Banerjee R, Ellis PD, Quail MA, Swerdlow HP, Zernicka-Goetz M, Livesey FJ, Ponting CP, Voet T (2015) G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat Methods 12(6):519–522
Angermueller C, Clark SJ, Lee HJ, Macaulay IC, Teng MJ, Hu TX, Krueger F, Smallwood SA, Ponting CP, Voet T, Kelsey G, Stegle O, Reik W (2016) Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat Methods 13(3):229–232
Hou Y, Guo H, Cao C, Li X, Hu B, Zhu P, Wu X, Wen L, Tang F, Huang Y, Peng J (2016) Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res 26(3):304–319
Li L, Guo F, Gao Y, Ren Y, Yuan P, Yan L, Li R, Lian Y, Li J, Hu B, Gao J, Wen L, Tang F, Qiao J (2018) Single-cell multi-omics sequencing of human early embryos. Nat Cell Biol 20(7):847–858
Cusanovich DA et al (2015) Multiplex single cell profiling of chromatin accessibility by combinatorial cellular indexing. Science (New York, N.Y.) 348(6237):910–914
Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP, Chang HY, Greenleaf WJ (2015) Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523(7561):486–490
Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP, Olsen BN, Mumbach MR, Pierce SE, Corces MR, Shah P, Bell JC, Jhutty D, Nemec CM, Wang J, Wang L, Yin Y, Giresi PG, Chang ALS, Zheng GXY, Greenleaf WJ, Chang HY (2019) Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 37(8):925–936
Granja JM, Klemm S, McGinnis LM, Kathiria AS, Mezger A, Corces MR, Parks B, Gars E, Liedtke M, Zheng GXY, Chang HY, Majeti R, Greenleaf WJ (2019) Single-cell multiomic analysis identifies regulatory programs in mixed-phenotype acute leukemia. Nat Biotechnol 37(12):1458–1465
Chen S, Lake BB, Zhang K (2019) High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol 37(12):1452–1457
Bian S et al (2018) Single-cell multiomics sequencing and analyses of human colorectal cancer. Science (New York, N.Y.) 362(6418):1060–1063
Acknowledgments
We thank Xing Wang, Meixi Liu, Zhenwei Tang, and Bowen Li for their guidance in revising the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 81972943 and No. 81830097) and the Hunan Talent Young Investigator (No. 2019RS2012).
Author information
Authors and Affiliations
Contributions
Concept and design: Qianjin Lu, Haijing Wu; drafting of the manuscript: Mingming Zhao; revising of the manuscript: Qianjin Lu, Haijing Wu, Ming Zhao, Christopher Chang.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, M., Jiang, J., Zhao, M. et al. The Application of Single-Cell RNA Sequencing in Studies of Autoimmune Diseases: a Comprehensive Review. Clinic Rev Allerg Immunol 60, 68–86 (2021). https://doi.org/10.1007/s12016-020-08813-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-020-08813-6