Abstract
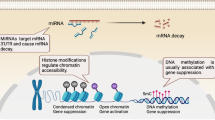
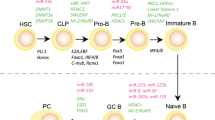
Autoreactive B cells are central in the pathogenesis of autoimmune diseases (AID) not only by producing autoantibodies but also by secreting cytokines and by presenting autoantigens. Changes in DNA methylation, histone modifications, and miRNA expression, the hallmarks of epigenetic failure, characterize B cells isolated from patients with AID, highlighting the contribution of epigenetic processes to autoreactivity. Additional evidence of epigenetic involvement in the development of B cell autoreactivity comes from in vivo and in vitro studies using DNA demethylating agents as accelerating factors or histone deacetylase inhibitors as repressing factors. As a result, a better understanding of the altered epigenetic processes in AID and in particular in B cells opens perspectives for the development of new therapeutics.

Similar content being viewed by others
Abbreviations
- AID:
-
Autoimmune diseases
- SLE:
-
Systemic lupus erythematosus
- RA:
-
Rheumatoid arthritis
- DNMT:
-
DNA methyltransferase
- HDAC:
-
Histone deacetylase
- miR:
-
MicroRNA
References
Hom G, Graham RR, Modrek B et al (2008) Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med 358:900–909
Fraga MF, Ballestar E, Paz MF et al (2005) Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci U S A 102:10604–10609
Garaud S, Le Dantec C, Jousse-Joulin S et al (2009) IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J Immunol 182:5623–5632
Balada E, Ordi-Ros J, Vilardell-Tarrés M (2007) DNA methylation and systemic lupus erythematosus. Ann N Y Acad Sci 1108:127–136
Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M (1990) Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum 33:1665–1673
Neidhart M, Rethage J, Kuchen S et al (2000) Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum 43:2634–2647
Lee YH, Rho YH, Choi SJ, Ji JD, Song GG (2007) PADI4 polymorphisms and rheumatoid arthritis susceptibility: a meta-analysis. Rheumatol Int 27:827–833
Webb R, Wren JD, Jeffries M et al (2009) Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum 60:1076–1084
Sawalha AH, Webb R, Han S et al (2008) Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS ONE 3:e1727
Chen CZ, Li L, Lodish HF, Bartel DP (2004) MicroRNAs modulate hematopoietic lineage differentiation. Science 303:83–86
Lindsay MA (2008) MicroRNAs and the immune response. Trends Immunol 29:343–351
Pauley KM, Satoh M, Chan AL, Bubb MR, Reeves WH, Chan EK (2008) Upregulated miR-146a expression in peripheral blood mononuclear cells from rheumatoid arthritis patients. Arthritis Res Ther 10:R101
Nakasa T, Miyaki S, Okubo A et al (2008) Expression of microRNA-146 in rheumatoid arthritis synovial tissue. Arthritis Rheum 58:1284–1292
Tang Y, Luo X, Cui H et al (2009) MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum 60:1065–1075
Dai Y, Huang YS, Tang M et al (2007) Microarray analysis of microRNA expression in peripheral blood cells of systemic lupus erythematosus patients. Lupus 16:939–946
Dai Y, Sui W, Lan H, Yan Q, Huang H, Huang Y (2008) Comprehensive analysis of microRNA expression patterns in renal biopsies of lupus nephritis patients. Rheumatol Int . doi:10.1007/s00296-008-0758-6
Danbara M, Kameyama K, Higashihara M, Takagaki Y (2002) DNA methylation dominates transcriptional silencing of Pax5 in terminally differentiated B cell lines. Mol Immunol 38:1161–1166
Amaravadi L, Klemsz MJ (1999) DNA methylation and chromatin structure regulate PU.1 expression. DNA Cell Biol 18:875–884
Maier H, Colbert J, Fitzsimmons D, Clark DR, Hagman J (2003) Activation of the early B-cell-specific mb-1 (Ig-alpha) gene by Pax-5 is dependent on an unmethylated Ets binding site. Mol Cell Biol 23:1946–1960
Walter K, Bonifer C, Tagoh H (2008) Stem cell-specific epigenetic priming and B cell-specific transcriptional activation at the mouse Cd19 locus. Blood 112:1673–1682
Schwab J, Illges H (2001) Silencing of CD21 expression in synovial lymphocytes is independent of methylation of the CD21 promoter CpG island. Rheumatol Int 20:133–137
O’Carroll D, Mecklenbrauker I, Das PP et al (2007) A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev 21:1999–2004
Koralov SB, Muljo SA, Galler GR et al (2008) Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132:860–874
Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD (2008) MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9:839–845
Thai TH, Calado DP, Casola S et al (2007) Regulation of the germinal center response by microRNA-155. Science 316:604–608
Rodriguez A, Vigorito E, Clare S et al (2007) Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611
Xiao C, Calado DP, Galler G et al (2007) MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159
Baxter J, Sauer S, Peters A et al (2004) Histone hypomethylation is an indicator of epigenetic plasticity in quiescent lymphocytes. EMBO J 23:4462–4472
Estève PO, Chin HG, Smallwood A et al (2006) Direct interaction between DNMT1 and G9a coordinates DNA and histone methylation during replication. Genes Dev 20:3089–3103
Thomas LR, Miyashita H, Cobb RM et al (2008) Functional analysis of histone methyltransferase g9a in B and T lymphocytes. J Immunol 181:485–493
Ehrlich M, Jackson K, Weemaes C (2006) Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF). Orphanet J Rare Dis 1:2
Blanco-Betancourt CE, Moncla A, Milili M et al (2004) Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood 103:2683–2690
Jin B, Tao Q, Peng J et al (2008) DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet 17:690–709
Ehrlich M, Sanchez C, Shao C et al (2008) ICF, an immunodeficiency syndrome: DNA methyltransferase 3B involvement, chromosome anomalies, and gene dysregulation. Autoimmunity 41:253–271
Lee SC, Bottaro A, Insel RA (2003) Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol Immunol 39:923–932
Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K (2000) Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol 20:2592–2603
Ancelin K, Lange UC, Hajkova P et al (2006) Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol 8:623–630
Gyory I, Wu J, Fejér G, Seto E, Wright KL (2004) PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat Immunol 5:299–308
Xu CR, Feeney AJ (2009) The epigenetic profile of Ig genes is dynamically regulated during B cell differentiation and is modulated by pre-B cell receptor signaling. J Immunol 182:1362–1369
Espinoza CR, Feeney AJ (2007) Chromatin accessibility and epigenetic modifications differ between frequently and infrequently rearranging VH genes. Mol Immunol 44:2675–2685
Nakase H, Takahama Y, Akamatsu Y (2003) Effect of CpG methylation on RAG1/RAG2 reactivity: implications of direct and indirect mechanisms for controlling V(D)J cleavage. EMBO 4:774–780
Larijani M, Frieder D, Sonbuchner TM et al (2005) Methylation protects cytidines from AID-mediated deamination. Mol Immunol 42:599–604
Odegard VH, Kim ST, Anderson SM, Shlomchik MJ, Schatz DG (2005) Histone modifications associated with somatic hypermutation. Immunity 23:101–110
Youinou P, Taher TE, Pers JO, Mageed RA, Renaudineau Y (2009) B-lymphocyte cytokines and rheumatic autoimmune disease. Arthritis Rheum 60:1873–1880
Harris DP, Haynes L, Sayles PC et al (2000) Reciprocal regulation of polarized cytokine production by effector B and T cells. Nat Immunol 1:475–482
Pang Y, Norihisa Y, Benjamin D, Kantor RR, Young HA (1992) Interferon-gamma gene expression in human B-cell lines: induction by interleukin-2, protein kinase C activators, and possible effect of hypomethylation on gene regulation. Blood 80:724–732
Janson PC, Marits P, Thörn M, Ohlsson R, Winqvist O (2008) CpG methylation of the IFNG gene as a mechanism to induce immunosuppression [correction of immunosupression] in tumor-infiltrating lymphocytes. J Immunol 181:2878–2886
Yamamoto I, Matsunaga T, Sakata K, Nakamura Y, Doi S, Hanmyou F (1996) Histone hyperacetylation plays a role in augmentation of IL-4-induced IgE production in LPS-stimulated murine B-lymphocytes by sodium butyrate. J Biochem 119:1056–1061
Cannat A, Seligmann M (1968) Induction by isoniazid and hydrallazine of antinuclear factors in mice. Clin Exp Immunol 3:99–105
Ten Veen JH, Feltkamp TE (1972) Studies on drug induced lupus erythematosus in mice. I. Drug induced antinuclear antibodies (ANA). Clin Exp Immunol 11:265–276
Dubroff LM, Reid RJ Jr (1980) Hydralazine-pyrimidine interactions may explain hydralazine-induced lupus erythematosus. Sciences 208:404–406
Lee BH, Yegnasubramanian S, Lin X, Nelson WG (2005) Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem 280:40749–40756
Deng C, Lu Q, Zhang Z et al (2003) Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum 48:746–756
Quddus J, Johnson KJ, Gavalchin J et al (1993) Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest 92:38–53
Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC (1995) Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol 154:3025–3035
Mazari L, Ouarzane M, Zouali M (2007) Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc Natl Acad Sci U S A 104:6317–6322
Bauer VW, Squire TL, Lowe ME, Andrews MT (2001) Expression of a chimeric retroviral-lipase mRNA confers enhanced lipolysis in a hibernating mammal. Am J Physiol Regul Integr Comp Physiol 281:R1186–R1192
Wu J, Zhou T, He J, Mountz JD (1993) Autoimmune disease in mice due to integration of an endogenous retrovirus in an apoptosis gene. J Exp Med 178:461–468
Qian Y, Santiago C, Borrero M, Tedder TF, Clarke SH (2001) Lupus-specific antiribonucleoprotein B cell tolerance in nonautoimmune mice is maintained by differentiation to B-1 and governed by B cell receptor signaling thresholds. J Immunol 166:2412–2419
Hippen KL, Tze LE, Behrens TW (2000) CD5 maintains tolerance in anergic B cells. J Exp Med 191:883–890
Jamin C, Magadur G, Lamour A et al (1992) Cell-free CD5 in patients with rheumatic diseases. Immunol Lett 31:79–83
Lu X, Axtell RC, Collawn JF, Gibson A, Justement LB, Raman C (2002) AP2 adaptor complex-dependent internalization of CD5: differential regulation in T and B cells. J Immunol 168:5612–5620
Renaudineau Y, Hillion S, Saraux A, Mageed RA, Youinou P (2005) An alternative exon 1 of the CD5 gene regulates CD5 expression in human B lymphocytes. Blood 106:2781–2789
Garaud S, Le Dantec C, Berthou C, Lydyard PM, Youinou P, Renaudineau Y (2008) Selection of the alternative exon 1 from the cd5 gene down-regulates membrane level of the protein in B lymphocytes. J Immunol 181:2010–2018
Renaudineau Y, Vallet S, Le Dantec C, Hillion S, Saraux A, Youinou P (2005) Characterization of the human CD5 endogenous retrovirus-E in B lymphocytes. Genes Immun 6:663–671
Pullmann R Jr, Bonilla E, Phillips PE, Middleton FA, Perl A (2008) Haplotypes of the HRES-1 endogenous retrovirus are associated with development and disease manifestations of systemic lupus erythematosus. Arthritis Rheum 58:532–540
Perl A, Colombo E, Dai H et al (1995) Antibody reactivity to the HRES-1 endogenous retroviral element identifies a subset of patients with systemic lupus erythematosus and overlap syndromes. Correlation with antinuclear antibodies and HLA class II alleles. Arthritis Rheum 38:1660–1671
Nagy G, Ward J, Mosser DD et al (2006) Regulation of CD4 expression via recycling by HRES-1/RAB4 controls susceptibility to HIV infection. J Biol Chem 281:34574–34591
Fernandez DR, Telarico T, Bonilla E et al (2009) Activation of mammalian target of rapamycin controls the loss of TCRzeta in lupus T cells through HRES-1/Rab4-regulated lysosomal degradation. J Immunol 182:2063–2073
Adelman MK, Marchalonis JJ (2002) Endogenous retroviruses in systemic lupus erythematosus: candidate lupus viruses. Clin Immunol 102:107–116
Chambers AE, Banerjee S, Chaplin T et al (2003) Histone acetylation-mediated regulation of genes in leukaemic cells. Eur J Cancer 39:1165–1175
Zabel MD, Weis JJ, Weis JH (1999) Lymphoid transcription of the murine CD21 gene is positively regulated by histone acetylation. J Immunol 163:2697–2703
Seo JS, Cho NY, Kim HR et al (2008) Cell cycle arrest and lytic induction of EBV-transformed B lymphoblastoid cells by a histone deacetylase inhibitor, Trichostatin A. Oncol Rep 19:93–98
Mishra N, Reilly CM, Brown DR, Ruiz P, Gilkeson GS (2003) Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J Clin Invest 111:539–552
Xiao C, Srinivasan L, Calado DP et al (2008) Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol 9:405–414
Park BL, Kim LH, Shin HD, Park YW, Uhm WS, Bae SC (2004) Association analyses of DNA methyltransferase-1 (DNMT1) polymorphisms with systemic lupus erythematosus. J Hum Genet 49:642–646
Gorelik G, Fang JY, Wu A, Sawalha AH, Richardson B (2007) Impaired T cell protein kinase C delta activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J Immunol 179:5553–5563
Miyamoto A, Nakayama K, Imaki H et al (2002) Increased proliferation of B cells and auto-immunity in mice lacking protein kinase Cdelta. Nature 416:865–869
Jury EC, Isenberg DA, Mauri C, Ehrenstein MR (2006) Atorvastatin restores Lck expression and lipid raft-associated signaling in T cells from patients with systemic lupus erythematosus. J Immunol 177:7416–7422
Oelke K, Lu Q, Richardson D et al (2004) Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum 50:1850–1860
Nagumo H, Agematsu K (1998) Synergistic augmentative effect of interleukin-10 and CD27/CD70 interactions on B-cell immunoglobulin synthesis. Immunology 94:388–394
Liang B, Gardner DB, Griswold DE, Bugelski PJ, Song XY (2006) Anti-interleukin-6 monoclonal antibody inhibits autoimmune responses in a murine model of systemic lupus erythematosus. Immunology 119:296–305
Tang LP, Cho CH, Hui WM et al (2006) An inverse correlation between Interleukin-6 and select gene promoter methylation in patients with gastric cancer. Digestion 74:85–90
Hillion S, Garaud S, Devauchelle V et al (2007) Interleukin-6 is responsible for aberrant B-cell receptor-mediated regulation of RAG expression in systemic lupus erythematosus. Immunology 122:371–380
Hu N, Qiu X, Luo Y et al (2008) Abnormal histone modification patterns in lupus CD4+ T cells. J Rheumatol 35:804–810
Lei W, Luo Y, Yan K et al (2009) Abnormal DNA methylation in CD4+ T cells from patients with systemic lupus erythematosus, systemic sclerosis, and dermatomyositis. Scand J Rheumatol 14:1–6
Huber LC, Brock M, Hemmatazad H et al (2007) Histone deacetylase/acetylase activity in total synovial tissue derived from rheumatoid arthritis and osteoarthritis patients. Arthritis Rheum 56:1087–1093
Acknowledgement
Thanks are due to Dr. WH Brooks (Moffitt Research Institute, Tampa, USA) for editorial assistance. We are also grateful to Cindy Séné and Simone Forest for their secretarial help.
Author information
Authors and Affiliations
Corresponding author
Additional information
This work was supported by grants from the Conseil Regional de Bretagne, the Conseil Général du Finistère, and the French Ministry for Education and Research.
Rights and permissions
About this article
Cite this article
Renaudineau, Y., Garaud, S., Le Dantec, C. et al. Autoreactive B Cells and Epigenetics. Clinic Rev Allerg Immunol 39, 85–94 (2010). https://doi.org/10.1007/s12016-009-8174-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12016-009-8174-6