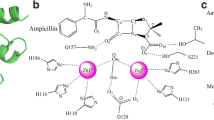
Abstract
Antimicrobial resistance is an emerging threat to public health around the world. The study employs computational and biophysical methods to investigate the properties of cefotaxime and meropenem’s binding to various beta-lactamases like TEM-1, SHV-1, KPC-2, and Amp-C. The enzyme kinetics of purified proteins revealed an increase in Michaelis constant (Km) value in the presence of meropenem and cefotaxime, indicating a decrease in enzyme affinity for nitrocefin. Proteins interact with meropenem/cefotaxime, causing quenching through complex formation. All proteins have one binding site, and binding constant (Kb) values are 104, indicating strong interaction. The study found that meropenem and cefotaxime had high fitness scores for Amp-C, KPC-2,TEM-1 and SHV-1, with binding energy ranging from −7.4 to −7.8, and hydrogen bonds between them. Molecular Dynamic simulation of protein–ligand complexes revealed cefotaxime-binding proteins have slightly lower Root Mean Square Deviation(RMSD) than meropenem-binding proteins, indicating stable association antibiotics with these proteins.






Similar content being viewed by others
Data availability
No datasets were generated or analyzed during the current study.
References
Pulingam, T., Parumasivam, T., Gazzali, A. M., Sulaiman, A. M., Chee, J. Y., Lakshmanan, M., & Sudesh, K. (2022). Antimicrobial resistance: prevalence, economic burden, mechanisms of resistance and strategies to overcome. European Journal of Pharmaceutical Sciences, 170, 106103. https://doi.org/10.1016/j.ejps.2021.106103.
Papp-Wallace, K. M. (2019). The latest advances in β-lactam/β-lactamase inhibitor combinations for the treatment of Gram-negative bacterial infections. Expert Opinion on Pharmacotherapy, 20(17), 2169–2184. https://doi.org/10.1080/14656566.2019.1660772.
Pitout, J. D. D., Sanders, C. C., & Sanders, W. E. (1997). Antimicrobial resistance with focus on β-lactam resistance in gram-negative bacilli. The American Journal of Medicine, 103(1), 51–59. https://doi.org/10.1016/S0002-9343(97)00044-2.
Drawz, S. M., & Bonomo, R. A. (2010). Three decades of β-lactamase inhibitors. Clinical Microbiology Reviews, 23(1), 160–201. https://doi.org/10.1128/CMR.00037-09.
Bush, K., & Jacoby, G. A. (2010). Updated functional classification of β-lactamases. Antimicrobial Agents and Chemotherapy, 54(3), 969–976. https://doi.org/10.1128/AAC.01009-09.
Spratt, B. G., & Cromie, K. D. (1988). Penicillin-binding proteins of gram-negative bacteria. Clinical Infectious Diseases, 10(4), 699–711. https://doi.org/10.1093/clinids/10.4.699.
Oka, T., Hashizume, K., & Fujita, H. (1980). Inhibition of peptidoglycan transpeptidase by beta-lactam antibiotics: structure-activity relationships. The Journal of Antibiotics, 33(11), 1357–1362. https://doi.org/10.7164/antibiotics.33.1357.
Zeng, X., & Lin, J. (2013). Beta-lactamase induction and cell wall metabolism in Gram-negative bacteria. Frontiers in Microbiology, 4, 128.
Philippon, A.(1985). Susceptibility of Pseudomonas aeruginosa to beta-lactam antibiotics. Chemioterapia: International Journal of the Mediterranean Society of Chemotherapy, 4(6), 424–8.
Farhat, N., Gupta, D., Ali, A., Kumar, Y., Akhtar, F., Kulanthaivel, S., … Khan, A. U. (2022). Broad-spectrum inhibitors against class A, B, and C type β-lactamases to block the hydrolysis against antibiotics: kinetics and structural characterization. Microbiology Spectrum, 10(5), e0045022.
Kuzin, A. P., Nukaga, M., Nukaga, Y., Hujer, A. M., Bonomo, R. A., & Knox, J. R. (1999). Structure of the SHV-1 β-Lactamase. Biochemistry, 38(18), 5720–5727. https://doi.org/10.1021/bi990136d.
Fonzé, E., Charlier, P., To’th, Y., Vermeire, M., Raquet, X., Dubus, A., & Frère, J.-M. (1995). TEM1 β-lactamase structure solved by molecular replacement and refined structure of the S235A mutant. Acta Crystallographica Section D Biological Crystallography, 51(5), 682–694. https://doi.org/10.1107/S0907444994014496.
Petrella, S., Ziental-Gelus, N., Mayer, C., Renard, M., Jarlier, V., & Sougakoff, W. (2008). Genetic and structural insights into the dissemination potential of the extremely broad-spectrum class A β-Lactamase KPC-2 identified in an Escherichia coli strain and an enterobacter cloacae strain isolated from the same patient in france. Antimicrobial Agents and Chemotherapy, 52(10), 3725–3736. https://doi.org/10.1128/AAC.00163-08.
McKinney, D. C., Zhou, F., Eyermann, C. J., Ferguson, A. D., Prince, D. B., Breen, J., & Verheijen, J. C. (2015). 4,5-disubstituted 6-Aryloxy-1,3-dihydrobenzo[c][1,2]oxaboroles are broad-spectrum serine β-lactamase inhibitors. ACS Infectious Diseases, 1(7), 310–316. https://doi.org/10.1021/acsinfecdis.5b00031.
Farhat, N., & Khan, A. U. (2021). Repurposing drug molecule against SARS-Cov-2 (COVID-19) through molecular docking and dynamics: a quick approach to pick FDA-approved drugs. Journal of Molecular Modeling, 27(11), 312. https://doi.org/10.1007/s00894-021-04923-w.
Kim, S., Chen, J., Cheng, T., Gindulyte, A., He, J., He, S., & Bolton, E. E. (2019). Nucleic Acids Research, 47(D1), D1102–D1109.
Jones, G., Willett, P., & Glen, R. C. (1995). Molecular recognition of receptor sites using a genetic algorithm with a description of desolvation. Journal of Molecular Biology, 245(1), 43–53. https://doi.org/10.1016/S0022-2836(95)80037-9.
Trott, O., & Olson, A. J. (2009). AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. Journal of Computational Chemistry, 31, 455–461.
Dalal, V., & Kumari, R. (2022). Screening and Identification of Natural Product‐Like Compounds as Potential Antibacterial Agents Targeting FemC of Staphylococcus aureus: An in‐Silico Approach. ChemistrySelect, 7(42). https://doi.org/10.1002/slct.202201728.
Dalal, V., Dhankhar, P., Singh, V., Singh, V., Rakhaminov, G., Golemi-Kotra, D., & Kumar, P. (2021). Structure-based identification of potential drugs against FmtA of Staphylococcus aureus: virtual screening, molecular dynamics, MM-GBSA, and QM/MM. The Protein Journal, 40(2), 148–165. https://doi.org/10.1007/s10930-020-09953-6.
Kumari, R., & Dalal, V. (2022). Identification of potential inhibitors for LLM of Staphylococcus aureus: structure-based pharmacophore modeling, molecular dynamics, and binding free energy studies. Journal of Biomolecular Structure and Dynamics, 40(20), 9833–9847. https://doi.org/10.1080/07391102.2021.1936179.
Singh, V., Dhankhar, P., Dalal, V., Tomar, S., Golemi-Kotra, D., & Kumar, P. (2022). Drug-repurposing approach to combat Staphylococcus aureus: biomolecular and binding interaction study. ACS Omega, 7(43), 38448–38458. https://doi.org/10.1021/acsomega.2c03671.
Kumari, R., Rathi, R., Pathak, S. R., & Dalal, V. (2022). Structural-based virtual screening and identification of novel potent antimicrobial compounds against YsxC of Staphylococcus aureus. Journal of Molecular Structure, 1255, 132476 https://doi.org/10.1016/j.molstruc.2022.132476.
Qamar, M., Sultanat, S., Khan, A. U., Ali, A., & Farhat, N. (2022). One pot facile synthesis of flavanoidal oxadiazinanones: In vitro antibacterial activity, docking and MD simulation using DNA gyrase. Journal of Molecular Structure, 1251, 131944. https://doi.org/10.1016/j.molstruc.2021.131944.
Beg, A. Z., Farhat, N., & Khan, A. U. (2021). Designing multi-epitope vaccine candidates against functional amyloids in Pseudomonas aeruginosa through immunoinformatic and structural bioinformatics approach. Infection, Genetics and Evolution, 93, 104982. https://doi.org/10.1016/j.meegid.2021.104982.
Sharma, M., Farhat, N., Khan, A. U., Khan, F. H., & Mahmood, R. (2023). Studies on the interaction of 2,4-dibromophenol with human hemoglobin using multi-spectroscopic, molecular docking and molecular dynamics techniques. Journal of Biomolecular Structure and Dynamics, 1–11. https://doi.org/10.1080/07391102.2023.2264975.
Lahiri, S. D., Bradford, P. A., Nichols, W. W., & Alm, R. A. (2016). Structural and sequence analysis of class A β-lactamases with respect to avibactam inhibition: impact of Ω-loop variations. Journal of Antimicrobial Chemotherapy, 71(10), 2848–2855. https://doi.org/10.1093/jac/dkw248.
Sun, J., Chikunova, A., Boyle, A. L., Voskamp, P., Timmer, M., & Ubbink, M. (2023). Enhanced activity against a third-generation cephalosporin by destabilization of the active site of a class A beta-lactamase. International Journal of Biological Macromolecules, 250, 126160. https://doi.org/10.1016/j.ijbiomac.2023.126160.
Benitez, M. J., Company, M., & Jiménez, J. S. (1991). Kinetics of the reaction between 5,5′-dithiobis[2-nitrobenzoic acid] and the sulphydryl group in Zn2+-dependent β-lactamase II. International Journal of Biological Macromolecules, 13(6), 345–348. https://doi.org/10.1016/0141-8130(91)90016-N.
Company, M., Benitez, M. J., & Jiménez, J. S. (1991). Degradation of β-lactam antibiotics in the presence of Zn2+ and 2-mino-2-hydroxymethylpropane-1,3-diol (Tris). A hypothetical non-enzymic model of β-lactamases. International Journal of Biological Macromolecules, 13(4), 225–230. https://doi.org/10.1016/0141-8130(91)90077-8.
Bhattacharya, S., Junghare, V., Pandey, N. K., Ghosh, D., Patra, H., & Hazra, S. (2020). An insight into the complete biophysical and biochemical characterization of novel class A beta-lactamase (Bla1) from Bacillus anthracis. International Journal of Biological Macromolecules, 145, 510–526. https://doi.org/10.1016/j.ijbiomac.2019.12.136.
Eftink, M. R., & Ghiron, C. A. (1981). Fluorescence quenching studies with proteins. Analytical Biochemistry, 114(2), 199–227. https://doi.org/10.1016/0003-2697(81)90474-7.
Farhat, N., Ali, A., Waheed, M., Gupta, D., & Khan, A. U. (2022). Chemically synthesised flavone and coumarin based isoxazole derivatives as broad spectrum inhibitors of serine β-lactamases and metallo-β-lactamases: a computational, biophysical and biochemical study. Journal of Biomolecular Structure and Dynamics, 0(0), 1–11. https://doi.org/10.1080/07391102.2022.2099977.
Bhattacharya, S., Padhi, A. K., Junghare, V., Das, N., Ghosh, D., Roy, P., & Hazra, S. (2021). Understanding the molecular interactions of inhibitors against Bla1 beta-lactamase towards unraveling the mechanism of antimicrobial resistance. International Journal of Biological Macromolecules, 177, 337–350. https://doi.org/10.1016/j.ijbiomac.2021.02.069.
Farhat, N., & Khan, A. U. (2024). Inhibitors against New Delhi metallo-betalactamase-1 (NDM-1) and its variants endemic in Indian settings along with the laboratory functional gain mutant of NDM-1. European Journal of Clinical Microbiology & Infectious Diseases. https://doi.org/10.1007/s10096-024-04761-7.
Farhat, N., & Khan, A. U. (2023). Repurposing FDA approved drug molecules against A B C classes of β-lactamases: a computational biology and molecular dynamics simulations study. Journal of Biomolecular Structure and Dynamics, 1–15. https://doi.org/10.1080/07391102.2023.2276890.
Farhat, N., Mujahid, S., & Khan, A. U. (2024). Mechanistic approach of effective combination of antibiotics against clinical bacterial strains having New Delhi Metallo-beta-lactamase variants and functional gain laboratory mutant. Current Microbiology, 81(1), 41 https://doi.org/10.1007/s00284-023-03553-0.
Acknowledgements
The authors acknowledge the facility and support provided by the Department of Biotechnology, Ministry of science and technology, Government of India.
Funding
Funding:The authors acknowledge funding from the DBT, Government of India, grant no. BT/PR40148/BTIS/137/20/2021, Tata Innovation Fellowship, BT/HRD/TIF/09/04/2021-22 and NNP DBT GRANT: BT/PR40180/BTIS/137/59/2023. NF is also recipient of senior research fellow from mentioned project.
Author information
Authors and Affiliations
Contributions
NF, performed all experiments and wrote manuscript, TK, performed some of the experiments, SN performed bioinformatic studies AUK, conceived problem and guided the study provided resources and checked first draft of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Farhat, N., Khanam, T., Noor, S. et al. Structural insight into the binding mode of cefotaxime and meropenem to TEM-1, SHV-1, KPC-2, and Amp-C type beta-lactamases. Cell Biochem Biophys (2024). https://doi.org/10.1007/s12013-024-01284-y
Accepted:
Published:
DOI: https://doi.org/10.1007/s12013-024-01284-y