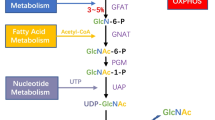
Abstract
O-GlcNAcylation, a recently discovered post-translational modification of proteins, plays a crucial role in regulating protein structure and function, and is closely associated with multiple diseases. Research has shown that O-GlcNAcylation is abnormally upregulated in most cancers, promoting disease progression. To elucidate the roles of O-GlcNAcylation in cancer, this review summarizes various cancer-associated biological events regulated by O-GlcNAcylation and the corresponding signaling pathways. This work may provide insights for future studies on the function or underlying mechanisms of O-GlcNAcylation in cancer.

Similar content being viewed by others
References
Xue, J., Zhu, L. P., & Wei, Q. (2013). IgG-Fc N-glycosylation at Asn297 and IgA O-glycosylation in the hinge region in health and disease. Glycoconjugate Journal, 30, 735–745. https://doi.org/10.1007/s10719-013-9481-y.
Thomas, D., Rathinavel, A. K. & Radhakrishnan, P. (2021). Altered glycosylation in cancer: a promising target for biomarkers and therapeutics. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer, 1875, 188464. https://doi.org/10.1016/j.bbcan.2020.188464.
Torres, C. R. & Hart, G. W. (1984). Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. Journal of of Biological Chemistry, 259, 3308–3317.
Issad, T., Masson, E., & Pagesy, P. (2010). O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes & Metabolism, 36, 423–435. https://doi.org/10.1016/j.diabet.2010.09.001.
Chatham, J. C., Zhang, J., & Wende, A. R. (2021). Role of O-Linked N-Acetylglucosamine protein modification in cellular (Patho)Physiology. Physiological Reviews, 101, 427–493. https://doi.org/10.1152/physrev.00043.2019.
Chan, J. J., Zhang, B., & Chew, X. H., et al. (2022). Pan-cancer pervasive upregulation of 3’ UTR splicing drives tumourigenesis. Nature Cell Biology, 24, 928–939. https://doi.org/10.1038/s41556-022-00913-z.
Sun, L., Zhang, H., & Gao, P. (2022). Metabolic reprogramming and epigenetic modifications on the path to cancer. Protein & Cell, 13, 877–919. https://doi.org/10.1007/s13238-021-00846-7.
East, M. P. & Johnson, G. L. (2022). Adaptive chromatin remodeling and transcriptional changes of the functional kinome in tumor cells in response to targeted kinase inhibition. Journal of Biological Chemistry, 298, 101525. https://doi.org/10.1016/j.jbc.2021.101525.
Lee, J. B., Pyo, K. H. & Kim, H. R. (2021). Role and function of O-GlcNAcylation in cancer. Cancers, 13. https://doi.org/10.3390/cancers13215365.
Hart, G. W., Slawson, C., & Ramirez-Correa, G., et al. (2011). Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry, 80, 825–858. https://doi.org/10.1146/annurev-biochem-060608-102511.
Chen, L., Zhou, Q., & Zhang, P., et al. (2023). Direct stimulation of de novo nucleotide synthesis by O-GlcNAcylation. Nature Chemical Biology. https://doi.org/10.1038/s41589-023-01354-x.
Shafi, R., Iyer, S. P., & Ellies, L. G., et al. (2000). The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proceedings of the National Academy of Sciences of USA, 97, 5735–5739. https://doi.org/10.1073/pnas.100471497.
Iyer, S. P., & Hart, G. W. (2003). Roles of the tetratricopeptide repeat domain in O-GlcNAc transferase targeting and protein substrate specificity. Journal of Biological Chemistry, 278, 24608–24616. https://doi.org/10.1074/jbc.M300036200.
Blatch, G. L., & Lassle, M. (1999). The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. Bioessays, 21, 932–939. 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N.
Wrabl, J. O., & Grishin, N. V. (2001). Homology between O-linked GlcNAc transferases and proteins of the glycogen phosphorylase superfamily. Journal Molecular Biology, 314, 365–374. https://doi.org/10.1006/jmbi.2001.5151.
Roos, M. D., & Hanover, J. A. (2000). Structure of O-linked GlcNAc transferase: mediator of glycan-dependent signaling. Biochemical and Biophysical Research Communications, 271, 275–280. https://doi.org/10.1006/bbrc.2000.2600.
Levine, Z. G., & Walker, S. (2016). The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annual Review of Biochemistry, 85, 631–657. https://doi.org/10.1146/annurev-biochem-060713-035344.
Trapannone, R., Mariappa, D., & Ferenbach, A. T., et al. (2016). Nucleocytoplasmic human O-GlcNAc transferase is sufficient for O-GlcNAcylation of mitochondrial proteins. Biochemical Journal, 473, 1693–1702. https://doi.org/10.1042/BCJ20160092.
Varshney, S., & Stanley, P. (2017). EOGT and O-GlcNAc on secreted and membrane proteins. Biochemical Society Transactions, 45, 401–408. https://doi.org/10.1042/BST20160165.
Barua, R., Mizuno, K., & Tashima, Y., et al. (2021). Bioinformatics and functional analyses implicate potential roles for EOGT and L-fringe in pancreatic cancers. Molecules, 26. https://doi.org/10.3390/molecules26040882.
Overdijk, B., Van der Kroef, W. M., & Van Steijn, G. J., et al. (1981). Isolation and further characterization of bovine brain hexosaminidase C. Biochimica et Biophysica Acta, 659, 255–266. https://doi.org/10.1016/0005-2744(81)90052-8.
King, D. T., Males, A., & Davies, G. J., et al. (2019). Molecular mechanisms regulating O-linked N-acetylglucosamine (O-GlcNAc)-processing enzymes. Current Opinion in Chemical Biology, 53, 131–144. https://doi.org/10.1016/j.cbpa.2019.09.001.
Ferron, M., Denis, M., & Persello, A., et al. (2018). Protein O-GlcNAcylation in cardiac pathologies: past, present, future. Frontiers in Endocrinology, 9, 819 https://doi.org/10.3389/fendo.2018.00819.
Marshall, S., Bacote, V., & Traxinger, R. R. (1991). Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. Journal of Biological Chemistry, 266, 4706–4712.
Sharma, N. S., Saluja, A. K., & Banerjee, S. (2018). “Nutrient-sensing” and self-renewal: O-GlcNAc in a new role. Journal of Bioenergetics and Biomembrances, 50, 205–211. https://doi.org/10.1007/s10863-017-9735-7.
Kopanja, D., Chand, V., & O’Brien, E., et al. (2022). Transcriptional repression by FoxM1 suppresses tumor differentiation and promotes metastasis of breast cancer. Cancer Research, 82, 2458–2471. https://doi.org/10.1158/0008-5472.CAN-22-0410.
Inoue, Y., Moriwaki, K., & Ueda, Y., et al. (2018). Elevated O-GlcNAcylation stabilizes FOXM1 by its reduced degradation through GSK-3beta inactivation in a human gastric carcinoma cell line, MKN45 cells. Biochemical and Biophysical Research Communications, 495, 1681–1687. https://doi.org/10.1016/j.bbrc.2017.11.179.
Caldwell, S. A., Jackson, S. R., & Shahriari, K. S., et al. (2010). Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene, 29, 2831–2842. https://doi.org/10.1038/onc.2010.41.
Grusche, F. A., Richardson, H. E., & Harvey, K. F. (2010). Upstream regulation of the hippo size control pathway. Current Biology, 20, R574–R582. https://doi.org/10.1016/j.cub.2010.05.023.
Halder, G., & Johnson, R. L. (2011). Hippo signaling: growth control and beyond. Development, 138, 9–22. https://doi.org/10.1242/dev.045500.
Moroishi, T., Hayashi, T., & Pan, W. W., et al. (2016). The Hippo pathway kinases LATS1/2 suppress cancer immunity. Cell, 167, 1525–1539. https://doi.org/10.1016/j.cell.2016.11.005.
Pan, D. (2010). The Hippo signaling pathway in development and cancer. Development Cell, 19, 491–505. https://doi.org/10.1016/j.devcel.2010.09.011.
Misra, J. R., & Irvine, K. D. (2018). The Hippo signaling network and its biological functions. Annual Review of Genetics, 52, 65–87. https://doi.org/10.1146/annurev-genet-120417-031621.
Hong, A. W., Meng, Z., & Guan, K. L. (2016). The Hippo pathway in intestinal regeneration and disease. Nature Reviews Gastroenterology & Hepatology, 13, 324–337. https://doi.org/10.1038/nrgastro.2016.59.
Peng, C., Zhu, Y., & Zhang, W., et al. (2017). Regulation of the Hippo-YAP pathway by glucose sensor O-GlcNAcylation. Molecular Cell, 68, 591–604. https://doi.org/10.1016/j.molcel.2017.10.010.
Xu, T. H., Sheng, Z., & Li, Y., et al. (2020). OGT knockdown counteracts high phosphate-induced vascular calcification in chronic kidney disease through autophagy activation by downregulating YAP. Life Science, 261, 118121 https://doi.org/10.1016/j.lfs.2020.118121.
Guo, H., Zhang, B., & Nairn, A. V., et al. (2017). O-Linked N-Acetylglucosamine (O-GlcNAc) expression levels epigenetically regulate colon cancer tumorigenesis by affecting the cancer stem cell compartment via modulating expression of transcriptional factor MYBL1. Journal of Biological Chemistry, 292, 4123–4137. https://doi.org/10.1074/jbc.M116.763201.
Lee, Y., & Rio, D. C. (2015). Mechanisms and regulation of alternative pre-mRNA splicing. Annual Review of Biochemistry, 84, 291–323. https://doi.org/10.1146/annurev-biochem-060614-034316.
Wu, S., & Näär, A. M. (2019). SREBP1-dependent de novo fatty acid synthesis gene expression is elevated in malignant melanoma and represents a cellular survival trait. Scientific Reports Uk, 9, 10369 https://doi.org/10.1038/s41598-019-46594-x.
Shimano, H., & Sato, R. (2017). SREBP-regulated lipid metabolism: convergent physiology - divergent pathophysiology. Nature Reviews Endocrinology, 13, 710–730. https://doi.org/10.1038/nrendo.2017.91.
Lee, G., Zheng, Y., & Cho, S., et al. (2017). Post-transcriptional regulation of De Novo Lipogenesis by mTORC1-S6K1-SRPK2 signaling. Cell, 171, 1545–1558. https://doi.org/10.1016/j.cell.2017.10.037.
Wang, H. Y., Lin, W., & Dyck, J. A., et al. (1998). SRPK2: a differentially expressed SR protein-specific kinase involved in mediating the interaction and localization of pre-mRNA splicing factors in mammalian cells. Journal of Cell Biology, 140, 737–750. https://doi.org/10.1083/jcb.140.4.737.
Jang, S. W., Liu, X., & Fu, H., et al. (2009). Interaction of Akt-phosphorylated SRPK2 with 14-3-3 mediates cell cycle and cell death in neurons. Journal of Biological Chemistry, 284, 24512–24525. https://doi.org/10.1074/jbc.M109.026237.
Tan, W., Jiang, P., & Zhang, W., et al. (2021). Posttranscriptional regulation of de novo lipogenesis by glucose-induced O-GlcNAcylation. Molecular Cell, 81, 1890–1904. https://doi.org/10.1016/j.molcel.2021.02.009.
Kohtz, J. D., Jamison, S. F., & Will, C. L., et al. (1994). Protein-protein interactions and 5’-splice-site recognition in mammalian mRNA precursors. Nature, 368, 119–124. https://doi.org/10.1038/368119a0.
Ferrer, C. M., Lynch, T. P., & Sodi, V. L., et al. (2014). O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Molecular Cell, 54, 820–831. https://doi.org/10.1016/j.molcel.2014.04.026.
Yi, W., Clark, P. M., & Mason, D. E., et al. (2012). Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science, 337, 975–980. https://doi.org/10.1126/science.1222278.
Rao, X., Duan, X., & Mao, W., et al. (2015). O-GlcNAcylation of G6PD promotes the pentose phosphate pathway and tumor growth. Nature Communications, 6, 8468 https://doi.org/10.1038/ncomms9468.
Clark, P. M., Dweck, J. F. & Mason, D. E. et al. (2008). Direct in-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. Journal of the American Chemical Society, 130, 11576–11577. https://doi.org/10.1021/ja8030467.
Park, J., Han, D., & Kim, K., et al. (2009). O-GlcNAcylation disrupts glyceraldehyde-3-phosphate dehydrogenase homo-tetramer formation and mediates its nuclear translocation. Biochimica et Biophysica Acta, 1794, 254–262. https://doi.org/10.1016/j.bbapap.2008.10.003.
Sirover, M. A. (2005). New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. Journal of Cellular Biochemistry, 95, 45–52. https://doi.org/10.1002/jcb.20399.
Wang, Y., Liu, J., & Jin, X., et al. (2017). O-GlcNAcylation destabilizes the active tetrameric PKM2 to promote the Warburg effect. Proceedings of the National Academy of Sciences USA, 114, 13732–13737. https://doi.org/10.1073/pnas.1704145115.
Koppenol, W. H., Bounds, P. L., & Dang, C. V. (2011). Otto Warburg’s contributions to current concepts of cancer metabolism. Nature Reviews Cancer, 11, 325–337. https://doi.org/10.1038/nrc3038.
Chen, Y., Bei, J., & Liu, M., et al. (2021). Sublethal heat stress-induced O-GlcNAcylation coordinates the Warburg effect to promote hepatocellular carcinoma recurrence and metastasis after thermal ablation. Cancer Letters, 518, 23–34. https://doi.org/10.1016/j.canlet.2021.06.001.
Song, H., Ma, J., & Bian, Z., et al. (2019). Global profiling of O-GlcNAcylated and/or phosphorylated proteins in hepatoblastoma. Signal Transduction and Targeted Therapy, 4, 40 https://doi.org/10.1038/s41392-019-0067-4.
Butkinaree, C., Park, K., & Hart, G. W. (2010). O-linked beta-N-acetylglucosamine (O-GlcNAc): Extensive crosstalk with phosphorylation to regulate signaling and transcription in response to nutrients and stress. Biochimica et Biophysica Acta, 1800, 96–106. https://doi.org/10.1016/j.bbagen.2009.07.018.
Hart, G. W., Housley, M. P., & Slawson, C. (2007). Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature, 446, 1017–1022. https://doi.org/10.1038/nature05815.
Wang, Z., Gucek, M., & Hart, G. W. (2008). Cross-talk between GlcNAcylation and phosphorylation: site-specific phosphorylation dynamics in response to globally elevated O-GlcNAc. Proceedingss of the National Academy of Sciences USA, 105, 13793–13798. https://doi.org/10.1073/pnas.0806216105.
Huang, H., Wu, Q., & Guo, X., et al. (2021). O-GlcNAcylation promotes the migratory ability of hepatocellular carcinoma cells via regulating FOXA2 stability and transcriptional activity. Journal of Cellular Physiology, 236, 7491–7503. https://doi.org/10.1002/jcp.30385.
Mendonsa, A. M., Na, T. Y., & Gumbiner, B. M. (2018). E-cadherin in contact inhibition and cancer. Oncogene, 37, 4769–4780. https://doi.org/10.1038/s41388-018-0304-2.
Hanover, J. A., Krause, M. W., & Love, D. C. (2012). Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nature Reviews Molecular Cell Biology, 13, 312–321. https://doi.org/10.1038/nrm3334.
Sakabe, K., Wang, Z., & Hart, G. W. (2010). Beta-N-acetylglucosamine (O-GlcNAc) is part of the histone code. Proceedings of the National Academy of Sciences USA, 107, 19915–19920. https://doi.org/10.1073/pnas.1009023107.
Fujiki, R., Chikanishi, T., & Hashiba, W., et al. (2009). GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature, 459, 455–459. https://doi.org/10.1038/nature07954.
Xie, X., Wu, Q. & Zhang, K. et al. (2021). O-GlcNAc modification regulates MTA1 transcriptional activity during breast cancer cell genotoxic adaptation. Biochimica et Biophysica Acta (BBA)-General Subjects, 1865, 129930. https://doi.org/10.1016/j.bbagen.2021.129930.
Pitt, J. M., Marabelle, A., & Eggermont, A., et al. (2016). Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Annals of Oncology, 27, 1482–1492. https://doi.org/10.1093/annonc/mdw168.
Burhol, P. G., Waldum, H. L. & Jorde, R. (1981). File and registration systems in a gastroenterological laboratory. Tidsskrift for den Norske Laegeforening: Tidsskrift for Praktisk Medicin, ny Raekke, 101, 1767–1770.
Qiang, A., Slawson, C., & Fields, P. E. (2021). The role of O-GlcNAcylation in immune cell activation. Frontiers in Endocrinology, 12, 596617 https://doi.org/10.3389/fendo.2021.596617.
Shapouri-Moghaddam, A., Mohammadian, S., & Vazini, H., et al. (2018). Macrophage plasticity, polarization, and function in health and disease. Journal of Cellular Physiology, 233, 6425–6440. https://doi.org/10.1002/jcp.26429.
Chang, Y. H., Weng, C. L., & Lin, K. I. (2020). O-GlcNAcylation and its role in the immune system. Journal of Biomedical Science, 27, 57 https://doi.org/10.1186/s12929-020-00648-9.
Rodrigues, M. N., Stanczak, M. A., & Oliveira, I. A., et al. (2020). Hyperglycemia enhances cancer immune evasion by inducing alternative macrophage polarization through increased O-GlcNAcylation. Cancer Immunology Research, 8, 1262–1272. https://doi.org/10.1158/2326-6066.CIR-19-0904.
Ouyang, M., Yu, C., & Deng, X., et al. (2022). O-GlcNAcylation and its role in cancer-associated inflammation. Frontiers in Immunology, 13, 861559 https://doi.org/10.3389/fimmu.2022.861559.
Li, X., Zhang, Z., & Li, L., et al. (2017). Myeloid-derived cullin 3 promotes STAT3 phosphorylation by inhibiting OGT expression and protects against intestinal inflammation. Journal of Experimental Medicine, 214, 1093–1109. https://doi.org/10.1084/jem.20161105.
Sung, H., Ferlay, J., & Siegel, R. L., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca: A Cancer Journal for Clinicians, 71, 209–249. https://doi.org/10.3322/caac.21660.
Taparra, K., Wang, H., & Malek, R., et al. (2018). O-GlcNAcylation is required for mutant KRAS-induced lung tumorigenesis. Journal of Clinical Investigation, 128, 4924–4937. https://doi.org/10.1172/JCI94844.
Tran, P. T., Fan, A. C., & Bendapudi, P. K., et al. (2008). Combined Inactivation of MYC and K-Ras oncogenes reverses tumorigenesis in lung adenocarcinomas and lymphomas. PloS One, 3, e2125 https://doi.org/10.1371/journal.pone.0002125.
Podsypanina, K., Politi, K., & Beverly, L. J., et al. (2008). Oncogene cooperation in tumor maintenance and tumor recurrence in mouse mammary tumors induced by Myc and mutant Kras. Proceedings of the National Academy of Sciences USA, 105, 5242–5247. https://doi.org/10.1073/pnas.0801197105.
Chou, T. Y., Hart, G. W., & Dang, C. V. (1995). c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. Journal of Biological Chemistry, 270, 18961–18965. https://doi.org/10.1074/jbc.270.32.18961.
Ilkhanizadeh, S., Lau, J., & Huang, M., et al. (2014). Glial progenitors as targets for transformation in glioma. Advances in Cancer Research, 121, 1–65. https://doi.org/10.1016/B978-0-12-800249-0.00001-9.
Buckner, J. C., Brown, P. D., & O’Neill, B. P., et al. (2007). Central nervous system tumors. Mayo Clinic Proceedings, 82, 1271–1286. https://doi.org/10.4065/82.10.1271.
Ciraku, L., Bacigalupa, Z. A., & Ju, J., et al. (2022). O-GlcNAc transferase regulates glioblastoma acetate metabolism via regulation of CDK5-dependent ACSS2 phosphorylation. Oncogene, 41, 2122–2136. https://doi.org/10.1038/s41388-022-02237-6.
Dang, C. V. (2012). Links between metabolism and cancer. Genes & Development, 26, 877–890. https://doi.org/10.1101/gad.189365.112.
Simic, P., Williams, E. O., & Bell, E. L., et al. (2013). SIRT1 suppresses the epithelial-to-mesenchymal transition in cancer metastasis and organ fibrosis. Cell Reports, 3, 1175–1186. https://doi.org/10.1016/j.celrep.2013.03.019.
Tao, T., He, Z., & Shao, Z., et al. (2016). TAB3 O-GlcNAcylation promotes metastasis of triple negative breast cancer. Oncotarget, 7, 22807–22818. https://doi.org/10.18632/oncotarget.8182.
Jia, C., Li, H., & Fu, D., et al. (2020). GFAT1/HBP/O-GlcNAcylation axis regulates β-catenin activity to promote pancreatic cancer aggressiveness. BioMed Research International, 2020, 1921609 https://doi.org/10.1155/2020/1921609.
Yang, C., Hu, J. F., & Zhan, Q., et al. (2021). SHCBP1 interacting with EOGT enhances O-GlcNAcylation of NOTCH1 and promotes the development of pancreatic cancer. Genomics, 113, 827–842. https://doi.org/10.1016/j.ygeno.2021.01.010.
Jiang, Y., Liu, Z., & Xu, F., et al. (2018). Aberrant O-glycosylation contributes to tumorigenesis in human colorectal cancer. Journal of Cellular and Molecular Medicine, 22, 4875–4885. https://doi.org/10.1111/jcmm.13752.
Orntoft, T. F., Harving, N., & Langkilde, N. C. (1990). O-linked mucin-type glycoproteins in normal and malignant colon mucosa: lack of T-antigen expression and accumulation of Tn and sialosyl-Tn antigens in carcinomas. International Journal of Cancer, 45, 666–672. https://doi.org/10.1002/ijc.2910450416.
Itzkowitz, S. H., Yuan, M., & Montgomery, C. K., et al. (1989). Expression of Tn, sialosyl-Tn, and T antigens in human colon cancer. Cancer Research, 49, 197–204.
Konno, A., Hoshino, Y., & Terashima, S., et al. (2002). Carbohydrate expression profile of colorectal cancer cells is relevant to metastatic pattern and prognosis. Clinical and Experimental Metastasis, 19, 61–70. https://doi.org/10.1023/a:1013879702702.
Schumacher, U., Higgs, D., & Loizidou, M., et al. (1994). Helix pomatia agglutinin binding is a useful prognostic indicator in colorectal carcinoma. Cancer, 74, 3104–3107. 10.1002/1097-0142(19941215)74:12<3104::aid-cncr2820741207>3.0.co;2-0.
Mi, W., Gu, Y., & Han, C., et al. (2011). O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochimica et Biophysica Acta, 1812, 514–519. https://doi.org/10.1016/j.bbadis.2011.01.009.
Liu, Y., & Peng, F. X. (2021). Research progress on O-GlcNAcylation in the occurrence, development, and treatment of colorectal cancer. World Journal Gastrointestinal Surgery, 13, 96–115. https://doi.org/10.4240/wjgs.v13.i2.96.
Castellano, G., Torrisi, E., & Ligresti, G., et al. (2009). The involvement of the transcription factor Yin Yang 1 in cancer development and progression. Cell Cycle, 8, 1367–1372. https://doi.org/10.4161/cc.8.9.8314.
Zhu, G., Qian, M., & Lu, L., et al. (2019) O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis. https://doi.org/10.1093/carcin/bgz010.
Zhang, X., Ma, L., & Qi, J., et al. (2015). MAPK/ERK signaling pathway-induced hyper-O-GlcNAcylation enhances cancer malignancy. Molecular and Cellular Biochemistry, 410, 101–110. https://doi.org/10.1007/s11010-015-2542-8.
Nie, H., Ju, H., & Fan, J., et al. (2020). O-GlcNAcylation of PGK1 coordinates glycolysis and TCA cycle to promote tumor growth. Nature Communications, 11, 36 https://doi.org/10.1038/s41467-019-13601-8.
Yu, M., Chu, S., & Fei, B., et al. (2019). O-GlcNAcylation of ITGA5 facilitates the occurrence and development of colorectal cancer. Experimental Cell Research, 382, 111464 https://doi.org/10.1016/j.yexcr.2019.06.009.
Gu, Y., Lu, K., & Yang, G., et al. (2014). Mutation spectrum of six genes in Chinese phenylketonuria patients obtained through next-generation sequencing. Plos One, 9, e94100 https://doi.org/10.1371/journal.pone.0094100.
Forshew, T., Murtaza, M., & Parkinson, C., et al. (2012). Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Science Translational Medicine, 4, 136r–168r. https://doi.org/10.1126/scitranslmed.3003726.
Kim, K. H., & Roberts, C. W. (2016). Targeting EZH2 in cancer. Nature Medicine, 22, 128–134. https://doi.org/10.1038/nm.4036.
Zheng, F., Liao, Y. J., & Cai, M. Y., et al. (2015). Systemic delivery of microRNA-101 potently inhibits hepatocellular carcinoma in vivo by repressing multiple targets. PloS Genetics, 11, e1004873 https://doi.org/10.1371/journal.pgen.1004873.
Tiwari, N., Tiwari, V. K., & Waldmeier, L., et al. (2013). Sox4 is a master regulator of epithelial-mesenchymal transition by controlling Ezh2 expression and epigenetic reprogramming. Cancer Cell, 23, 768–783. https://doi.org/10.1016/j.ccr.2013.04.020.
Tsanou, E., Peschos, D., & Batistatou, A., et al. (2008). The E-cadherin adhesion molecule and colorectal cancer. A global literature approach. Anticancer Research, 28, 3815–3826.
Becker, K. F., Rosivatz, E., & Blechschmidt, K., et al. (2007). Analysis of the E-Cadherin repressor snail in primary human cancers. Cells Tissues Organs, 185, 204–212. https://doi.org/10.1159/000101321.
Roy, H. K., Smyrk, T. C., & Koetsier, J., et al. (2005). The transcriptional repressor SNAIL is overexpressed in human colon cancer. Digestive Diseases and Sciences, 50, 42–46. https://doi.org/10.1007/s10620-005-1275-z.
Li, J., Ahmad, M., & Sang, L., et al. (2023). O-GlcNAcylation promotes the cytosolic localization of the m6A reader YTHDF1 and colorectal cancer tumorigenesis. Journal of Biological Chemistry, 104738. https://doi.org/10.1016/j.jbc.2023.104738.
Funding
This research was supported by National Natural Science Foundation of China (NSFC NO. 82102841 and NSFC NO. 82273094).
Author information
Authors and Affiliations
Contributions
Z.R. drafted the manuscript. M.D., L.Z., and Y.Z. provided suggestions and revised the manuscript. L.C. and Q.S. conceptualized, wrote, and revised the manuscript. And all authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ran, Z., Zhang, L., Dong, M. et al. O-GlcNAcylation: A Crucial Regulator in Cancer-Associated Biological Events. Cell Biochem Biophys 81, 383–394 (2023). https://doi.org/10.1007/s12013-023-01146-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-023-01146-z