Abstract
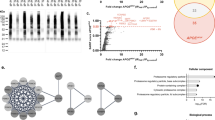
To understand adenosylmethionine decarboxylase 1 (AMD1)-mediated mRNA processing and cell adhesion activated & inhibited transition mechanisms between chimpanzee and human left hemisphere, AMD1-activated different complete (all no positive correlation, Pearson correlation coefficient < 0.25) and uncomplete (partly no positive correlation except AMD1, Pearson < 0.25) networks were identified in higher human compared with lower chimpanzee left hemisphere from the corresponding AMD1-stimulated (Pearson ≥ 0.25) or inhibited (Pearson ≤ −0.25) overlapping molecules of Pearson and GRNInfer, respectively. This result was verified by the corresponding scatter matrix. As visualized by GO, KEGG, GenMAPP, BioCarta, and disease database integration, we proposed mainly that AMD1-stimulated different complete network was involved in AMD1 activation with cytoplasm ubiquitin specific peptidase (tRNA-guanine transglycosylase) to nucleus paired box-induced mRNA processing, whereas the corresponding inhibited network participated in AMD1 repression with cytoplasm protocadherin gamma and adaptor-related protein complex 3-induced cell adhesion in lower chimpanzee left hemisphere. However, AMD1-stimulated network contained AMD1 activation with plakophilin and phosphodiesterase to SH3 binding glutamic acid-rich protein to dynein and zinc finger-induced cell adhesion, whereas the corresponding inhibited different complete network included AMD1 repression with mitochondrial denine nucleotide translocator, brain protein, and ADH dehydrogenase to ribonucleoprotein-induced mRNA processing in higher human left hemisphere. Our AMD1 different networks were verified by AMD1-activated or -inhibited complete and uncomplete networks within and between chimpanzee left hemisphere or (and) human left hemisphere.




Similar content being viewed by others
References
van Poelje, P. D., & Snell, E. E. (1990). Pyruvoyl-dependent enzymes. Annual Review of Biochemistry, 59, 29–59.
Pegg, A. E., Xiong, H., Feith, D. J., et al. (1998). S-adenosylmethionine decarboxylase: structure, function and regulation by polyamines. Biochemical Society Transactions, 26(4), 580–586.
Storey, J. D. (2002). A direct approach to false discovery rates. Journal of the Royal Statistical Society: Series B, 64, 479–498.
Wang, Y., Joshi, T., Zhang, X. S., et al. (2006). Inferring gene regulatory networks from multiple microarray datasets. Bioinformatics, 22(19), 2413–2420.
Wang, L., Huang, J., Jiang, M., et al. (2012). Activated PTHLH coupling feedback phosphoinositide to G-protein receptor signal-induced cell adhesion network in human hepatocellular carcinoma by systems-theoretic analysis. Scientific World Journal, 2012, 428979.
Wang, L., Huang, J., Jiang, M., et al. (2012). Inhibited PTHLH downstream leukocyte adhesion-mediated protein amino acid N-linked glycosylation coupling Notch and JAK-STAT cascade to iron-sulfur cluster assembly-induced aging network in no-tumor hepatitis/cirrhotic tissues (HBV or HCV infection) by systems-theoretical analysis. Integrative Biology (Camb), 4(10), 1256–1262.
Wang, L., Huang, J., Jiang, M., et al. (2012). Tissue-specific transplantation antigen P35B (TSTA3) immune response-mediated metabolism coupling cell cycle to postreplication repair network in no-tumor hepatitis/cirrhotic tissues (HBV or HCV infection) by biocomputation. Immunologic Research, 52(3), 258–268.
Wang, L., Huang, J., Jiang, M., et al. (2012). Signal transducer and activator of transcription 2 (STAT2) metabolism coupling postmitotic outgrowth to visual and sound perception network in human left cerebrum by biocomputation. Journal of Molecular Neuroscience, 47(3), 649–658.
Lin, H., Wang, L., Jiang, M., et al. (2012). P-glycoprotein (ABCB1) inhibited network of mitochondrion transport along microtubule and BMP signal-induced cell shape in chimpanzee left cerebrum by systems-theoretical analysis. Cell Biochemistry and Function, 30(7), 582–587.
Huang, J., Wang, L., Jiang, M., et al. (2012). PTHLH coupling upstream negative regulation of fatty acid biosynthesis and Wnt receptor signal to downstream peptidase activity-induced apoptosis network in human hepatocellular carcinoma by systems-theoretical analysis. Journal of Receptors and Signal Transduction Research, 32(5), 250–256.
Wang, L., Sun, L., Huang, J., et al. (2011). Cyclin-dependent kinase inhibitor 3 (CDKN3) novel cell cycle computational network between human non-malignancy associated hepatitis/cirrhosis and hepatocellular carcinoma (HCC) transformation. Cell Proliferation, 44(3), 291–299.
Wang, L., Huang, J., Jiang, M., et al. (2011). AFP computational secreted network construction and analysis between human hepatocellular carcinoma (HCC) and no-tumor hepatitis/cirrhotic liver tissues. Tumour Biology, 31(5), 417–425.
Wang, L., Huang, J., Jiang, M., et al. (2011). Survivin (BIRC5) cell cycle computational network in human no-tumor hepatitis/cirrhosis and hepatocellular carcinoma transformation. Journal of Cellular Biochemistry, 112(5), 1286–1294.
Wang, L., Huang, J., Jiang, M., et al. (2011). MYBPC1 computational phosphoprotein network construction and analysis between frontal cortex of HIV encephalitis (HIVE) and HIVE-control patients. Cellular and Molecular Neurobiology, 31(2), 233–241.
Wang, L., Huang, J., & Jiang, M. (2011). CREB5 computational regulation network construction and analysis between frontal cortex of HIV encephalitis (HIVE) and HIVE-control patients. Cell Biochemistry and Biophysics, 60(3), 199–207.
Wang, L., Huang, J., & Jiang, M. (2011). RRM2 computational phosphoprotein network construction and analysis between no-tumor hepatitis/cirrhotic liver tissues and human hepatocellular carcinoma (HCC). Cellular Physiology and Biochemistry, 26(3), 303–310.
Sun, L., Wang, L., Jiang, M., et al. (2011). Glycogen debranching enzyme 6 (AGL), enolase 1 (ENOSF1), ectonucleotide pyrophosphatase 2 (ENPP2_1), glutathione S-transferase 3 (GSTM3_3) and mannosidase (MAN2B2) metabolism computational network analysis between chimpanzee and human left cerebrum. Cell Biochemistry and Biophysics, 61(3), 493–505.
Huang, J. X., Wang, L., & Jiang, M. H. (2011). TNFRSF11B computational development network construction and analysis between frontal cortex of HIV encephalitis (HIVE) and HIVE-control patients. Journal of Inflammation (London), 7, 50.
Sun, Y., Wang, L., Jiang, M., et al. (2010). Secreted phosphoprotein 1 upstream invasive network construction and analysis of lung adenocarcinoma compared with human normal adjacent tissues by integrative biocomputation. Cell Biochemistry and Biophysics, 56(2–3), 59–71.
Huang, J., Wang, L., Jiang, M., et al. (2010). Interferon α-inducible protein 27 computational network construction and comparison between the frontal cortex of HIV encephalitis (HIVE) and HIVE-control patients. The Open Genomics Journal, 3, 1–8.
Wang, L., Sun, Y., Jiang, M., et al. (2009). Integrative decomposition procedure and Kappa statistics for the distinguished single molecular network construction and analysis. J Biomed Biotechnol, 2009, 726728.
Wang, L., Sun, Y., Jiang, M., et al. (2009). FOS proliferating network construction in early colorectal cancer (CRC) based on integrative significant function cluster and inferring analysis. Cancer Investigation, 27(8), 816–824.
Puente, X. S., Sanchez, L. M., Overall, C. M., et al. (2003). Human and mouse proteases: a comparative genomic approach. Nature Reviews Genetics, 4(7), 544–558.
Entrez Gene: USP14 ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase).
Mines, M. A., Goodwin, J. S., Limbird, L. E., et al. (2009). Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK activation. Journal of Biological Chemistry, 284(9), 5742–5752.
Lynch, E. D., Lee, M. K., Morrow, J. E., et al. (1997). Nonsyndromic deafness DFNA1 associated with mutation of a human homolog of the Drosophila gene diaphanous. Science, 278(5341), 1315–1318.
Sano, K., Tanihara, H., Heimark, R. L., et al. (1993). Protocadherins: a large family of cadherin-related molecules in central nervous system. EMBO Journal, 12(6), 2249–2256.
Entrez Gene: PCDHGC3 protocadherin gamma subfamily C, 3.
DuBois, R. N., McLane, M. W., Ryder, K., et al. (1990). A growth factor-inducible nuclear protein with a novel cysteine/histidine repetitive sequence. Journal of Biological Chemistry, 265(31), 19185–19191.
Schneller, D., Machat, G., Sousek, A., et al. (2011). p19(ARF)/p14(ARF) controls oncogenic functions of signal transducer and activator of transcription 3 in hepatocellular carcinoma. Hepatology, 54(1), 164–172.
Sanduja, S., Blanco, F. F., & Dixon, D. A. (2011). The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA, 2(1), 42–57.
Carballo, E., Lai, W. S., & Blackshear, P. J. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science, 281(5379), 1001–1005.
Johnson, B. A., Stehn, J. R., Yaffe, M. B., et al. (2002). Cytoplasmic localization of tristetraprolin involves 14-3-3-dependent and -independent mechanisms. Journal of Biological Chemistry, 277(20), 18029–18036.
Carman, J. A., & Nadler, S. G. (2004). Direct association of tristetraprolin with the nucleoporin CAN/Nup214. Biochemical and Biophysical Research Communications, 315(2), 445–449.
Myer, V. E., & Steitz, J. A. (1995). Isolation and characterization of a novel, low abundance hnRNP protein: A0. RNA, 1(2), 171–182.
Entrez Gene: HNRPA0 heterogeneous nuclear ribonucleoprotein A0.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 61171114), the State Key Laboratory of Drug Research (SIMM1302KF), Automatical Scientific Planning of Tsinghua University (20111081023 and 20111081010).
Conflict of interests
We have no conflict of interests.
Author information
Authors and Affiliations
Corresponding author
Additional information
Lin Wang, Juxiang Huang and Minghu Jiang have contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, L., Huang, J., Jiang, M. et al. Adenosylmethionine Decarboxylase 1 (AMD1)-Mediated mRNA Processing and Cell Adhesion Activated & Inhibited Transition Mechanisms by Different Comparisons Between Chimpanzee and Human Left Hemisphere. Cell Biochem Biophys 70, 279–288 (2014). https://doi.org/10.1007/s12013-014-9902-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12013-014-9902-y