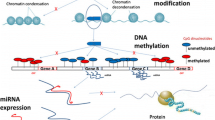
Abstract
Metal cadmium (Cd) and its compounds are ubiquitous industrial and environmental pollutants and they have been believed to exert severe damage to multiple organs and tissues. MicroRNAs (miRNAs) and long noncoding RNAs (lncRNAs) are the two most common noncoding RNAs and have pivotal roles in various cellular and physiological processes. Since the importance of miRNAs and lncRNAs in Cd toxicity has been widely recognized, we focus our interests on the current researches of miRNAs and lncRNAs as well as their regulation roles in Cd toxicity. In this paper, the keywords “cadmium” in combination with “miRNA” or “LncRNA” or “noncoding RNA” was used to retrieve relevant articles in PubMed, EMbase, CNKI, Wan Fang, and CBM databases. The literatures which contained the above keywords and carried out in animals (in vivo and in vitro) have been collected, collated, analyzed, and summarized. Our summary results showed that hundreds of miRNAs and lncRNAs are involved in the Cd toxicity, which have been demonstrated as multiple organ injury, reproductive toxicity, malignant transformation, and abnormal repair of DNA damage. In this paper, we also discussed the blank in present research field of Cd toxicity as well as suggested some ideas for future study in Cd toxicity.



Similar content being viewed by others
References
ATSDR’s Substance Priority List. Available online: https://www.atsdr.cdc.gov/spl/index.html (accessed on 30 January 2018)
Lyon. France Beryllium, cadmium, mercury and exposures in the glass manufacturing industry. International Agency for Research on Cancer, 58, 119–238 (IARC 1993)
Peralta-Videa JR, Lopez ML, Narayan M, Saupe G, Gardea-Torresdey J (2009) The biochemistry of environmental heavy metal uptake by plants: implications for the food chain. Int J Biochem Cell Biol 41:1665–1677
Ghosh S, Rabha R, Chowdhury M, Padhy PK (2018) Source and chemical species characterization of PM10 and human health risk assessment of semi-urban, urban and industrial areas of West Bengal, India. Chemosphere 207:626–636
Thévenod F, Lee WK (2013) Toxicology of cadmium and its damage to mammalian organs. Met Ions Life Sci 11:415–490
Larsson SC, Wolk A (2016) Urinary cadmium and mortality from all causes, cancer and cardiovascular disease in the general population: systematic review and meta-analysis of cohort studies. Int J Epidemiol 45:782–791
Lv Y, Wang P, Huang R, Liang X, Wang P, Tan J, Chen Z, Dun Z, Wang J, Jiang Q, Wu S, Ling H, Li Z, Yang X (2017) Cadmium exposure and osteoporosis: a population-based study and benchmark dose estimation in southern China. J Bone Miner Res 32:1990–2000
Da Cunha Martins A Jr, Carneiro MF, Grotto D, Adeyemi JA, Barbosa F Jr (2018) Arsenic, cadmium, and mercury-induced hypertension: mechanisms and epidemiological findings. J Toxicol Environ Health B Crit Rev 21:61–82
Rinaldi M, Micali A, Marini H, Adamo EB, Puzzolo D, Pisani A, Trichilo V, Altavilla D, Squadrito F, Minutoli L (2017) Cadmium, organ toxicity and therapeutic approaches: a review on brain, kidney and testis damage. Curr Med Chem 24:3879–3893
Liu W, Zhang B, Huang Z, Pan X, Chen X, Hu C, Liu H, Jiang Y, Sun X, Peng Y, Xia W, Xu S, Li Y (2018) Cadmium body burden and gestational diabetes mellitus: a prospective study. Environ Health Perspect 126:027006
Byber K, Lison D, Verougstraete V, Dressel H, Hotz P (2016) Cadmium or cadmium compounds and chronic kidney disease in workers and the general population: a systematic review. Crit Rev Toxicol 46:191–240
Gao M, Li C, Xu M, Liu Y, Cong M, Liu S (2018) LncRNA MT1DP aggravates cadmium-induced oxidative stress by repressing the function of Nrf2 and is dependent on interaction with miR-365. Adv Sci (Weinh) 5:1800087
Jin X, Jia T, Liu R, Xu S (2018) The antagonistic effect of selenium on cadmium-induced apoptosis via PPAR-γ/PI3K/Akt pathway in chicken pancreas. J Hazard Mater 357:355–362
Fujishiro H, Liu Y, Ahmadi B, Templeton DM (2018) Protective effect of cadmium-induced autophagy in rat renal mesangial cells. Arch Toxicol 92:619–631
Cowley M, Skaar DA, Jima DD, Maguire RL, Hudson KM, Park SS, Sorrow P, Hoyo C (2018) Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and newborn children. Environ Health Perspect 126:037003
Fay MJ, Alt LAC, Ryba D, Salamah R, Peach R, Papaeliou A, Zawadzka S, Weiss A, Patel N, Rahman A, Stubbs-Russell Z, Lamar PC, Edwards JR, Prozialeck WC (2018) Cadmium nephrotoxicity is associated with altered microRNA expression in the rat renal cortex. Toxics 6:E16
Pellegrini KL, Gerlach CV, Craciun F (2016) Application of small RNA sequencing to identify microRNAs in acute kidney injury and fibrosis. Toxicol Appl Pharmacol 312:42–52
Chen Z, Gu D, Zhou M, Shi H, Yan S, Cai Y (2016) Regulatory role of miR-125a/b in the suppression by selenium of cadmium-induced apoptosis via the mitochondrial pathway in LLC-PK1 cells. Chem Biol Interact 243:35–44
Fabbri M, Urani C, Sacco MG, Procaccianti C, Gribaldo L (2012) Whole genome analysis and microRNAs regulation in HepG2 cells exposed to cadmium. ALTEX 29:173–182
Chen M, Li X, Fan R, Yang J, Jin X, Hamid S, Xu S (2018) Cadmium induces BNIP3-dependent autophagy in chicken spleen by modulating miR-33-AMPK axis. Chemosphere 194:396–402
Hassan F, Nuovo G, Crawford M, Boyaka P, Kirkby S, Nana-Sinkam S, Cormet-Boyaka E (2012) MiR-101 and miR-144 regulate the expression of the CFTR chloride channel in the lung. PLoS One 7:e50837
De-Ugarte L, Balcells S, Nogues X, Grinberg D, Diez-Perez A, Garcia-Giralt N (2018) Pro-osteoporotic miR-320a impairs osteoblast function and induces oxidative stress. PLoS One 13:e0208131
Bollati V, Marinelli B, Apostoli P, Bonzini M, Nordio F, Hoxha M, Pegoraro V, Motta V (2010) Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ Health Perspect 118:763–768
Wang W, Chen J, Luo L, Li Y, Liu J, Zhang W (2018) Effect of cadmium on kitl pre-mRNA alternative splicing in murine ovarian granulosa cells and its associated regulation by miRNAs[J]. J Appl Toxicol 38:227–239
Weng S, Wang W, Li Y, Li H, Lu X, Xiao S, Wu T, Xie M, Zhang W (2014) Continuous cadmium exposure from weaning to maturity induces downregulation of ovarian follicle development-related SCF/c-kit gene expression and the corresponding changes of DNA methylation/microRNA pattern. Toxicol Lett 225:367–377
Li Q, Kappil MA, Li A, Dassanayake PS, Darrah TH, Friedman AE, Friedman M, Lambertini L, Landrigan P (2015) Exploring the associations between microRNA expression profiles and environmental pollutants in human placenta from the National Children’s Study (NCS). Epigenetics 10:793–802
Brooks SA, Martin E, Smeester L, Grace MR, Boggess K, Fry RC (2016) miRNAs as common regulators of the transforming growth factor (TGF)-β pathway in the preeclamptic placenta and cadmium-treated trophoblasts: Links between the environment, the epigenome and preeclampsia. Food Chem Toxicol 98:50–57
Brooks SA, Fry RC (2017) Cadmium inhibits placental trophoblast cell migration via miRNA regulation of the transforming growth factor beta (TGF-β) pathway. Food Chem Toxicol 109:721–726
Hu H, Lu X, Cen X, Chen X, Li F, Zhong S (2014) RNA-Seq identifies key reproductive gene expression alterations in response to cadmium exposure. Biomed Res Int 2014:529271
Gao F, Zhang P, Zhang H, Zhang Y, Zhang Y, Hao Q, Zhang X (2017) Dysregulation of long noncoding RNAs in mouse testes and spermatozoa after exposure to cadmium. Biochem Biophys Res Commun 484:8–14
Liu Q, Zheng C, Shen H, Zhou Z, Lei Y (2015) MicroRNAs-mRNAs expression profile and their potential role in malignant transformation of human bronchial epithelial cells induced by cadmium. Biomed Res Int 2015:902025
Urani C, Melchioretto P, Bruschi M, Fabbri M, Sacco M, Gribaldo L (2015) Impact of cadmium on intracellular Zinc levels in HepG2 cells: quantitative evaluations and molecular effects. Biomed Res Int 2015:949514
Martínez-Pacheco M, Hidalgo-Miranda A, Romero-Córdoba S, Valverde M, Rojas E (2014) MRNA and miRNA expression patterns associated to pathways linked to metal mixture health effects. Gene. 533:508–514
Zhou Z, Liu H, Wang C, Lu Q, Huang Q, Zheng C, Lei Y (2015) Long non-coding RNAs as novel expression signatures modulate DNA damage and repair in cadmium toxicology. Sci Rep 5:15293
Tani H, Onuma Y, Ito Y, Torimura M (2014) Long non-coding RNAs as surrogate indicators for chemical stress responses in human-induced pluripotent stem cells. PLoS One 9:e106282
Bartel DP (2018) Metazoan microRNAs. Cell 173:20–51
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15:509–524
Nguyen TA, Jo MH, Choi YG, Park J, Kwon SC, Hohng S, Kim VN, Woo JS (2015) Functional anatomy of the human microprocessor. Cell 161:1374–1387
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Rådmark O, Kim S, Kim VN (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425:415–419
Lund E, Güttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303:95–98
Iwasaki S, Kobayashi M, Yoda M, Sakaguchi Y, Katsuma S, Suzuki T, Tomari Y (2010) Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol Cell 39:292–299
Wang S, Wu W, Claret FX (2017) Mutual regulation of microRNAs and DNA methylation in human cancers. Epigenetics 12:187–197
Alarcón CR, Lee H, Goodarzi H, Halberg N, Tavazoie SF (2015) N6-methyladenosine marks primary microRNAs for processing. Nature 519:482–485
Gu S, Sun D, Dai H, Zhang Z (2018) N6-methyladenosine mediates the cellular proliferation and apoptosis via microRNAs in arsenite-transformed cells. Toxicol Lett 292:1–11
Berulava T, Rahmann S, Rademacher K, Klein-Hitpass L, Horsthemke B (2015) N6-adenosine methylation in miRNAs. PLoS One 10:e0118438
Furió-Tarí P, Tarazona S, Gabaldón T, Enright AJ, Conesa A (2017) spongeScan: a web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res 44:W176–W180
Militello G, Weirick T, John D, Döring C, Dimmeler S, Uchida S (2017) Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform 18:780–788
Thomson DW, Dinger ME (2016) Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet 17:272–283
Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172:393–407
Kim C, Kang D, Lee EK, Lee JS (2017) Long noncoding RNAs and RNA-binding proteins in oxidative stress, cellular senescence, and age-related diseases. Oxidative Med Cell Longev 2017:2062384
Wu X, Tudoran OM, Calin GA, Ivan M (2018) The many faces of long noncoding RNAs in cancer[J]. Antioxid Redox Signal 29:922–935
Greco S, Salgado Somoza A, Devaux Y, Martelli F (2018) Long noncoding RNAs and cardiac disease. Antioxid Redox Signal 29:880–901
Zhou M, Zhao H, Wang X, Sun J, Su J (2018) Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief Bioinform 20:598–608
Losko M, Kotlinowski J, Jura J (2016) Long noncoding RNAs in metabolic syndrome related disorders. Mediat Inflamm 2016:5365209
Tang L, Shen H, Li X, Li Z, Liu Z, Xu J, Ma S, Zhao X, Bai X, Li M, Wang Q, Ji J (2016) MiR-125a-5p decreases after long non-coding RNA HOTAIR knockdown to promote cancer cell apoptosis by releasing caspase 2. Cell Death Dis 7:e2137
Lei L, Kang H, Guo J, Shi X, Zhang Z, Gao, Zhang G (2019) Renal impairment induced by environmental cadmium exposure: role of TGF-β1/Smad3 and related microRNAs. J Environ Occup Med 36:511–518
Xie Y, Chu A, Feng Y, Chen L, Shao Y, Luo Q, Deng X, Wu M, Shi X, Chen Y (2018) MicroRNA-146a: a comprehensive indicator of inflammation and oxidative stress status induced in the brain of chronic T2DM rats. Front Pharmacol 9:478
Liu KX, Chen GP, Lin PL, Huang JC, Lin X, Qi JC, Lin QC (2018) Detection and analysis of apoptosis- and autophagy-related miRNAs of mouse vascular endothelial cells in chronic intermittent hypoxia model. Life Sci 193:194–199
Brognara E, Fabbri E, Montagner G, Gasparello J, Manicardi A, Corradini R, Bianchi N, Finotti A, Breveglieri G, Borgatti M (2016) High levels of apoptosis are induced in human glioma cell lines by co-administration of peptide nucleic acids targeting miR-221 and miR-222. Int J Oncol 48:1029–1038
Gao M, Chen M, Li C, Xu M, Liu Y, Cong M, Sang N, Liu S (2018) Long non-coding RNA MT1DP shunts the cellular defense to cytotoxicity through crosstalk with MT1H and RhoC in cadmium stress. Cell Discov 4:5–24
Ji YL, Wang H, Zhao XF, Wang Q, Zhang C, Zhang Y, Zhao M, Chen YH, Meng XH, Xu DX (2011) Crosstalk between endoplasmic reticulum stress and mitochondrial pathway mediates cadmium-induced germ cell apoptosis in testes. Toxicol Sci 124:446–459
Guo FZ, Zhang LS, Wei JL (2016) Endosulfan inhibiting the meiosis process via depressing expressions of regulatory factors and causing cell cycle arrest in spermatogenic cells. Environ Sci Pollut Res Int 23:20506–20516
Funding
This review was supported by grant from the Dali University PhD Research Initiation Fund Project 2018 (KYBS2018006) and China Southwest Collaborative Innovation Center for Entomoceutics Open Project in 2018 (NO. CIC1802) to Shiyan GU.
Author information
Authors and Affiliations
Contributions
Shiyan-GU and Jiao-DAI wrote the manuscript. Shiyan Gu, Zuoshun-HE, and Tengjiao-QU revised the manuscript and checked the references.
Corresponding authors
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Gu, S., Dai, J., Qu, T. et al. Emerging Roles of MicroRNAs and Long Noncoding RNAs in Cadmium Toxicity. Biol Trace Elem Res 195, 481–490 (2020). https://doi.org/10.1007/s12011-019-01859-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12011-019-01859-4