Abstract
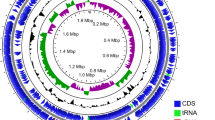
This study aims to sequence the whole genome of Pediococcus ethanolidurans CP201 isolated from Daqu and determine the anti-corrosion ability of bacteriocins on chicken breast. The whole genome sequence information of P. ethanolidurans CP201 was analyzed, and its gene structure and function were explored. It was found that gene1164 had annotations in the NR, Pfam, and Swiss-Prot databases, and was related to bacteriocins. The exogenous expression of the bacteriocin gene Pediocin PE-201 was analyzed based on the pET-21b vector and the host BL21, and the corresponding bacteriocin was successfully expressed under the induction of IPTG. After purification by NI–NTA column, enterokinase treatment, membrane dialysis concentration treatment, and SDS-PAGE electrophoresis, the molecular weight was about 6.5 kDa and the purity was above 90%. By applying different concentrations of bacteriocin to chicken breast with different levels of contamination, the control of pathogenic bacteria, the ordinary contamination level (OC) group, and the high contamination level (MC) group could be completely achieved with 25 mg/L bacteriocin. In conclusion, the bacteriocin produced by the newly isolated CP201 can be applied to the preservation of meat products to prevent the risk of food-borne diseases.





Similar content being viewed by others
Availability of Data and Materials
The author states that the data and other documents supporting the results of this research can be found in the article. In addition, the sequence data reported in this paper has been stored in NCBI SRA database (PRJNA884871).
References
CDC. (2021). Salmonella. Centers for disease control and prevention. Retrieved from https://www.cdc.gov/salmonella/index.html. Accessed 11 Oct 2021.
Li, Y., Yang, X., Zhang, H., Jia, H., & Yang, D. (2020). Prevalence and antimicrobial susceptibility of Salmonella in the commercial eggs in China. International Journal of Food Microbiology, 325, 108623.
Li, Q., Xin, W., Yin, K., Hu, Y., & Jiao, X. (2017). Genetic analysis and crispr typing of Salmonella enterica serovar enteritidis from different sources revealed potential transmission from poultry and pig to human. International Journal of Food Microbiology, 266, 119–125.
Souza, M. N., Lehmann, F. K. M., De Carli, S., Kipper, D., Fonseca, A. S. K., Ikuta, N., & Lunge, V. R. (2019). Molecular detection of Salmonella serovars enteritidis, heidelberg and typhimurium directly from pre-enriched poultry samples. British Poultry Science, 60(4), 388–394.
Guohan, M. A., Huanhuan, M. A., Xinran, L., Liu, J., Sun, Y., Bai, F., & Jianrong, L. (2019). Screening for broad-spectrum antagonistic lactic acid bacteria from intestine of turbot and identi? cation of bacteriocin produced by it. Food Science, 40(6), 159–165.
Yi, L., Luo, L., & Lü, X. (2018). Efficient exploitation of multiple novel bacteriocins by combination of complete genome and peptidome. Frontiers in Microbiology, 9, 1567.
Arbulu, S., Jiménez, J. J., Gútiez, L., Feito, J., Cintas, L. M., Herranz, C., & Hernández, P. E. (2019). Cloning and expression of synthetic genes encoding native, hybrid-and bacteriocin-derived chimeras from mature class IIa bacteriocins, by Pichia pastoris (syn. Komagataella spp.). Food Research International, 121, 888–899.
Cao, S., Du, R., Zhao, F., Xiao, H., Han, Y., & Zhou, Z. (2019). The mode of action of bacteriocin CHQS, a high antibacterial activity bacteriocin produced by Enterococcus faecalis TG2. Food Control, 96, 470–478.
Lv, X., Ma, H., Sun, M., Lin, Y., Bai, F., Li, J., & Zhang, B. (2018). A novel bacteriocin DY4-2 produced by Lactobacillus plantarum from cutlassfish and its application as bio-preservative for the control of Pseudomonas fluorescens in fresh turbot (Scophthalmus maximus) fillets. Food Control, 89, 22–31.
Deegan, L. H., Cotter, P. D., Hill, C., & Ross, P. (2006). Bacteriocins: Biological tools for bio-preservation and shelf-life extension. International Dairy Journal, 16(9), 1058–1071.
Gabrielsen, C., Brede, D. A., Nes, I. F., & Diep, D. B. (2014). Circular bacteriocins: Biosynthesis and mode of action. Applied and Environmental Microbiology, 80(22), 6854–6862.
Ghanbari, M., Jami, M., Domig, K. J., & Kneifel, W. (2013). Seafood biopreservation by lactic acid bacteria–A review. LWT-Food Science and Technology, 54(2), 315–324.
Calo-Mata, P., Arlindo, S., Boehme, K., de Miguel, T., Pascoal, A., & Barros-Velazquez, J. (2008). Current applications and future trends of lactic acid bacteria and their bacteriocins for the biopreservation of aquatic food products. Food and Bioprocess Technology, 1(1), 43–63.
Pei, J., Li, X., Han, H., & Tao, Y. (2018). Purification and characterization of plantaricin SLG1, a novel bacteriocin produced by Lb. plantarum isolated from yak cheese. Food Control, 84, 111–117.
Martinez, F. A. C., Balciunas, E. M., Converti, A., Cotter, P. D., & de Souza Oliveira, R. P. (2013). Bacteriocin production by Bifidobacterium spp. A review. Biotechnology Advances, 31(4), 482–488.
Zheng, X. W., Tabrizi, M. R., Nout, M. R., & Han, B. Z. (2011). Daqu—A traditional Chinese liquor fermentation starter. Journal of the Institute of Brewing, 117(1), 82–90.
Hu, Y., Huang, X., Yang, B., Zhang, X., Han, Y., Chen, X. X., & Han, B. Z. (2021). Contrasting the microbial community and metabolic profile of three types of light-flavor Daqu. Food Bioscience, 44, 101395.
He, G., Huang, J., Zhou, R., Wu, C., & Jin, Y. (2019). Effect of fortified Daqu on the microbial community and flavor in Chinese strong-flavor liquor brewing process. Frontiers in Microbiology, 10, 56.
Zhang, L., Wu, C., Ding, X., Zheng, J., & Zhou, R. (2014). Characterisation of microbial communities in Chinese liquor fermentation starters Daqu using nested PCR-DGGE. World Journal of Microbiology and Biotechnology, 30(12), 3055–3063.
Tran, N. H., Rahman, M. Z., He, L., Xin, L., Shan, B., & Li, M. (2016). Complete de novo assembly of monoclonal antibody sequences. Scientific Reports, 6(1), 1–10.
Porto, W. F., Pires, A. S., & Franco, O. L. (2017). Computational tools for exploring sequence databases as a resource for antimicrobial peptides. Biotechnology Advances, 35(3), 337–349.
Queiroz, L. L., Hoffmann, C., Lacorte, G. A., de Melo Franco, B. D. G., & Todorov, S. D. (2022). Genomic and functional characterization of bacteriocinogenic lactic acid bacteria isolated from Boza, a traditional cereal-based beverage. Scientific Reports, 12(1), 1–13.
Rodriguez-R, L. M., Gunturu, S., Harvey, W. T., Rosselló-Mora, R., Tiedje, J. M., Cole, J. R., & Konstantinidis, K. T. (2018). The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Research, 46(W1), W282–W288.
Chan, P. P., Lin, B. Y., Mak, A. J., & Lowe, T. M. (2021). tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Research, 49(16), 9077–9096.
Bland, C., Ramsey, T. L., Sabree, F., Lowe, M., Brown, K., Kyrpides, N. C., & Hugenholtz, P. (2007). CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics, 8(1), 1–8.
Yella, V. R., Kumar, A., & Bansal, M. (2018). Identification of putative promoters in 48 eukaryotic genomes on the basis of DNA free energy. Scientific Reports, 8(1), 1–13.
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., & Kuipers, O. P. (2018). BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Research, 46(W1), W278–W281.
Moon, G. S., Pyun, Y. R., & Kim, W. J. (2006). Expression and purification of a fusion-typed pediocin PA-1 in Escherichia coli and recovery of biologically active pediocin PA-1. International Journal of Food Microbiology, 108(1), 136–140.
Beaulieu, L., Tolkatchev, D., Jette, J. F., Groleau, D., & Subirade, M. (2007). Production of active pediocin PA-1 in Escherichia coli using a thioredoxin gene fusion expression approach: Cloning, expression, purification, and characterization. Canadian Journal of Microbiology, 53(11), 1246–1258.
Xin, L. Ř, Yi, L., Dang, J., Dang, Y., & Liu, B. (2014). Purification of novel bacteriocin produced by Lactobacillus coryniformis MXJ 32 for inhibiting bacterial foodborne pathogens including antibiotic-resistant microorganisms. Food Control, 46, 264–271.
De Souza, G. T., De Carvalho, R. J., De Sousa, J. P., Tavares, J. F., Schaffner, D., De Souza, E. L., & Magnani, M. (2016). Effects of the essential oil from Origanum vulgare L. on survival of pathogenic bacteria and starter lactic acid bacteria in semihard cheese broth and slurry. Journal of Food Protection, 79(2), 246–252.
Nakano, C., Ozawa, H., Akanuma, G., Funa, N., & Horinouchi, S. (2009). Biosynthesis of aliphatic polyketides by type III polyketide synthase and methyltransferase in Bacillus subtilis. Journal of Bacteriology, 191(15), 4916–4923.
Seshime, Y., Juvvadi, P. R., Kitamoto, K., Ebizuka, Y., & Fujii, I. (2010). Identification of csypyrone B1 as the novel product of Aspergillus oryzae type III polyketide synthase CsyB. Bioorganic & Medicinal Chemistry, 18(12), 4542–4546.
Gillespie, D. E., Brady, S. F., Bettermann, A. D., Cianciotto, N. P., Liles, M. R., Rondon, M. R., & Handelsman, J. (2002). Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Applied and Environmental Microbiology, 68(9), 4301–4306.
Rattray, J. E., van de Vossenberg, J., Hopmans, E. C., Kartal, B., van Niftrik, L., Rijpstra, W. I. C., Strous, M., Jetten, M. S. M., Schouten, S., & Damsté, J. S. S. (2008). Ladderane lipid distribution in four genera of anammox bacteria. Archives of Microbiology, 190(1), 51–66.
Tietz, J. I., Schwalen, C. J., Patel, P. S., Maxson, T., & Blair, P. M. (2017). A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nature Chemical Biology, 13(5), 470–478.
Cap, M., Paredes, P. F., Fernández, D., Mozgovoj, M., Vaudagna, S. R., & Rodriguez, A. (2020). Effect of high hydrostatic pressure on Salmonella spp inactivation and meat-quality of frozen chicken breast. LWT, 118, 108873.
Albano, H., Todorov, S. D., van Reenen, C. A., Hogg, T., Dicks, L. M., & Teixeira, P. (2007). Characterization of two bacteriocins produced by Pediococcus acidilactici isolated from “Alheira”, a fermented sausage traditionally produced in Portugal. International Journal of Food Microbiology, 116(2), 239–247.
Loessner, M., Guenther, S., Steffan, S., & Scherer, S. (2003). A pediocin-producingsLactobacillus plantarum strain inhibits Listeria monocytogenes in a multispecies cheese surface microbial ripening consortium. Applied and Environmental Microbiology, 69(3), 1854–1857.
Osmanagaoglu, O., Kiran, F., & Nes, I. F. (2011). A probiotic bacterium, Pediococcus pentosaceus OZF, isolated from human breast milk produces pediocin AcH/PA-1. African Journal of Biotechnology, 10(11), 2070–2079.
Chen, Y., Ludescher, R. D., & Montville, T. J. (1998). Influence of lipid composition on pediocin PA-1 binding to phospholipid vesicles. Applied and Environmental Microbiology, 64(9), 3530–3532.
Santiago-Silva, P., Soares, N. F., Nóbrega, J. E., Júnior, M. A., Barbosa, K. B., Volp, A. C. P., Zerdas, E., & Würlitzer, N. J. (2009). Antimicrobial efficiency of film incorporated with pediocin (ALTA® 2351) on preservation of sliced ham. Food Control, 20(1), 85–89.
Ceruso, M., Liu, Y. H., Gunther, N. W., Pepe, T., Anastasio, A., Qi, P. X., Tomasula, P. M., & Renye, J. A. (2021). Anti-listerial activity of thermophilin 110 and pediocin in fermented milk and whey. Food Control, 125, 107941.
Berry, E. D., Hutkins, R. W., & Mandigo, R. W. (1991). The use of bacteriocin-producing Pediococcus acidilactici to control postprocessing Listeria monocytogenes contamination of frankfurters. Journal of Food Protection, 54(9), 681–686.
Meade, E., Slattery, M. A., & Garvey, M. (2020). Bacteriocins, potent antimicrobial peptides and the fight against multi drug resistant species: Resistance is futile? Antibiotics, 9(1), 32.
Funding
This work was supported by the National Natural Science Foundation of China (Project No. 30660005), the Sichuan Tujiu Liquor Co., Ltd (Project No. 222305), the Protect of Chengdu Shuzhiyuan Liquor Co., Ltd (Project No. 202306), the Protect of Chengdu Technology Innovation (Project No. 2022-YF05-00136-SN), and the Project of Sichuan Institute of International Science and Technology Cooperation (Australia and New Zealand) (Project No. AXYJ2022 -005).
Author information
Authors and Affiliations
Contributions
LZL: writing—original draft preparation; YL: methodology; ZP: writing—review and editing; LT: data curation, software, validation; ZJH: data curation; TWG: funding acquisition, writing—reviewing and editing. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
All authors declared their consent to participate.
Consent to Publish
All authors declare their consent to publish their work.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Guan, T., Long, L., Liu, Y. et al. Complete Genome Sequencing and Bacteriocin Functional Characterization of Pediococcus ethanolidurans CP201 from Daqu. Appl Biochem Biotechnol 195, 4728–4743 (2023). https://doi.org/10.1007/s12010-023-04575-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04575-x