Abstract
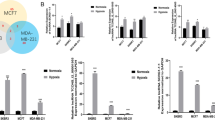
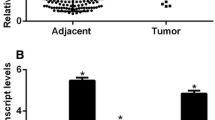
Breast cancer is the most malignant subtype of gynecological tumors and with aggressive biological behavior and poor outcome. Ultra-conserved non-coding RNA (ucRNA) is a newly discovered class of long non-coding RNAs (lncRNAs) which involved in the regulation of interaction network of genes. However, the exact function and mechanism by which ucRNA modulates breast cancer aggressive has not yet to be completely elucidated. In the present study, we demonstrated that the expression of uc.246 was significantly upregulated in metastatic breast cancer patients and TNBC cell lines, compared with those in controls. Furthermore, overexpression of uc.246 in MCF-7 cell lines enhanced the capacity of breast cancer cells to induce tube formation and migration of HUVECs, and, finally, enhanced breast cancer cells metastasis. Meanwhile, uc.246 overexpressing enhances the EMT phenotype of TNBC cells. Mechanistically, we found that uc.246 promoted malignant progression of breast cancer via upregulating the levels of VEGF-C and increased the levels of mesenchymal marker protein. Our results demonstrated that uc.246 induced angiogenesis, migration, and EMT phenotype and may represent a novel prognostic biomarker and therapeutic target for patients with breast cancer.






Similar content being viewed by others
Data Availability
All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
References
Sung, H., Ferlay, J., Siegel, R. L., et al. (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 71, 209–249.
Soerjomataram, I., & Bray, F. (2021). Planning for tomorrow: Global cancer incidence and the role of prevention 2020–2070. Nature Reviews Clinical Oncology, 18, 663–672.
Elston, C. W., Ellis, I. O., & Pinder, S. E. (1999). Pathological prognostic factors in breast cancer. Critical Reviews in Oncology/Hematology, 31, 209–223.
Hill, D. A., Friend, S., Lomo, L., et al. (2018). Breast cancer survival, survival disparities, and guideline-based treatment. Breast Cancer Research and Treatment, 170, 405–414.
Raica, M., Jitariu, A. A., & Cimpean, A. M. (2016). Lymphangiogenesis and anti-lymphangiogenesis in cutaneous melanoma. Anticancer Research, 36, 4427–4435.
Al-Rawi, M. A., & Jiang, W. G. (2011). Lymphangiogenesis and cancer metastasis. Frontiers in Bioscience, 16, 723–739.
Alitalo, K., Tammela, T., & Petrova, T. V. (2005). Lymphangiogenesis in development and human disease. Nature, 438, 946–953.
Mariotti, V., Fiorotto, R., Cadamuro, M., et al. (2021). New insights on the role of vascular endothelial growth factor in biliary pathophysiology. JHEP Reports: Innovation in Hepatology, 3, 100251.
McColl, B. K., Stacker, S. A., & Achen, M. G. (2004). Molecular regulation of the VEGF family – Inducers of angiogenesis and lymphangiogenesis. APMIS: Acta Pathologica, Microbiologica, et Immunologica Scandinavica, 112, 463–480.
Hu, X., & Luo, J. (2018). Heterogeneity of tumor lymphangiogenesis: Progress and prospects. Cancer Science, 109, 3005–3012.
Song, E., Mao, T., Dong, H., et al. (2020). VEGF-C-driven lymphatic drainage enables immunosurveillance of brain tumours. Nature, 577, 689–694.
Li, W. N., Hsiao, K. Y., Wang, C. A., et al. (2020). Extracellular vesicle-associated VEGF-C promotes lymphangiogenesis and immune cells infiltration in endometriosis. Proceedings of the National Academy of Sciences of the United States of America, 117, 25859–25868.
Karaman, S., & Detmar, M. (2014). Mechanisms of lymphatic metastasis. The Journal of Clinical Investigation, 124, 922–928.
Dieterich, L. C., & Detmar, M. (2016). Tumor lymphangiogenesis and new drug development. Advanced Drug Delivery Reviews, 99, 148–160.
Mittal, V. (2018). Epithelial mesenchymal transition in tumor metastasis. Annual Review of Pathology, 13, 395–412.
Zhang, Y., & Weinberg, R. A. (2018). Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Frontiers of Medicine, 12, 361–373.
Chen, T., You, Y., Jiang, H., et al. (2017). Epithelial-mesenchymal transition (EMT): A biological process in the development, stem cell differentiation, and tumorigenesis. Journal of Cellular Physiology, 232, 3261–3272.
Esteller, M. (2011). Non-coding RNAs in human disease. Nature Reviews Genetics, 12, 861–874.
Liz, J., Portela, A., Soler, M., et al. (2014). Regulation of pri-miRNA processing by a long noncoding RNA transcribed from an ultraconserved region. Molecular Cell, 55, 138–147.
Terracciano, D., Terreri, S., de Nigris, F., et al. (2017). The role of a new class of long noncoding RNAs transcribed from ultraconserved regions in cancer. Biochimica et Biophysica Acta Reviews on Cancer, 1868, 449–455.
Bejerano, G., Pheasant, M., Makunin, I., et al. (2004). Ultraconserved elements in the human genome. Science, 304, 1321–1325.
Zhou, J., Wang, R., Zhang, J., et al. (2017). Conserved expression of ultra-conserved noncoding RNA in mammalian nervous system. Biochimica et Biophysica Acta Gene Regulatory Mechanisms, 1860, 1159–1168.
Jiang, B. C., Yang, T., He, L. N., et al. (2016). Altered T-UCRs expression profile in the spinal cord of mice with neuropathic pain. Translational Perioperative and Pain Medicine, 1, 1–10.
Kopp, F., & Mendell, J. T. (2018). Functional classification and experimental dissection of long noncoding RNAs. Cell, 172, 393–407.
Wilusz, J. E., Sunwoo, H., & Spector, D. L. (2009). Long noncoding RNAs: Functional surprises from the RNA world. Genes & Development, 23, 1494–1504.
Hombach, S., & Kretz, M. (2016). Non-coding RNAs: Classification, biology and functioning. Advances in Experimental Medicine and Biology, 937, 3–17.
Malmuthuge, N., & Guan, L. L. (2021). Noncoding RNAs: Regulatory molecules of host-microbiome crosstalk. Trends in Microbiology, 29, 713–724.
Zhang, Y., Mao, Q., Xia, Q., et al. (2021). Noncoding RNAs link metabolic reprogramming to immune microenvironment in cancers. Journal of Hematology & Oncology, 14, 169.
Cunnick, G. H., Jiang, W. G., Gomez, K. F., et al. (2002). Lymphangiogenesis and breast cancer metastasis. Histology and Histopathology, 17, 863–870.
Roy, S., Banerjee, P., Ekser, B., et al. (2021). Targeting lymphangiogenesis and lymph node metastasis in liver cancer. The American Journal of Pathology, 191, 2052–2063.
Sammarco, G., Varricchi, G., Ferraro, V., et al. (2019). Mast cells, angiogenesis and lymphangiogenesis in human gastric cancer. International Journal of Molecular Sciences, 20(9), 2106.
Acknowledgements
Not applicable.
Funding
This work was in part supported by grants from the Natural Science Foundation of China (No.82003116).
Author information
Authors and Affiliations
Contributions
YL and XPH were performed experiments, analysis and interpretation of data, YL and WFX design and wrote manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no known competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Y., Hu, X. & Xia, W. Overexpression of Long Non-coding RNA uc.246 Facilitates Angiogenesis, Migration, and EMT Phenotype of Human Breast Cancer Cells. Appl Biochem Biotechnol 196, 1142–1153 (2024). https://doi.org/10.1007/s12010-023-04572-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04572-0