Abstract
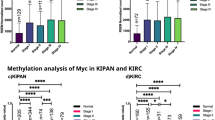
In this study, we analysed the Myc expression in the pan-kidney cohort (KIPAN) and kidney renal clear cell carcinoma (KIRC) in human tumour tissues compared to normal tissues. Myc is overexpressed and associated with poor overall survival (OS) in the KIPAN and KIRC. It shows that Myc plays a crucial role in the growth and maintenance of these malignancies. Additionally, we explored coexpressed genes, gene-set enrichment analysis of coexpressed genes, proteins and regulatory partners directly linked with Myc in KIPAN and KIRC and their role in cancer-specific events. Pathway enrichment analysis concluded that Myc-related genes are involved in many cancer-related pathways. Furthermore, we studied that among KIPAN, mutant forms of tumour suppressor genes have a poor prognosis and are associated with higher Myc expression but not in KIRC. This paper also investigates the correlation between Myc expression and promoter methylation, tumour-infiltrating lymphocytes, and the interaction of Myc with drugs. Our study indicates that Myc can be used as a diagnostic and prognostic biomarker in patients with KIPAN and KIRC with diverse clinical and pathological characteristics.
Graphical Abstract









Similar content being viewed by others
Data Availability
RNA sequencing data from the Cancer Genome Atlas (TCGA) was extracted from the Broad GDAC Firehose (http://gdac.broadinstitute.org/). Renal cancer datasets [KIPAN and KIRC] were used in the current study.
Abbreviations
- RCC:
-
Renal cell carcinoma
- KIPAN:
-
Pan-kidney cohort (KICH + KIRC + KIRP)
- KICH and chrRCC:
-
Kidney chromophobe
- KIRC and ccRCC:
-
Kidney renal clear cell carcinoma
- KIRP and pRCC:
-
Kidney renal papillary cell carcinoma
- OS:
-
Overall survival
- TP53:
-
Tumour protein 53
- VH:
-
Von Hippel-Lindau syndrome
- SETD2:
-
SET domain containing 2, histone lysine methyltransferase
- HIFs:
-
Hypoxia inducible factors
- TCGA:
-
The Cancer Genome Atlas
- TIMER:
-
Tumour immune estimation resource
- CTD:
-
Comparative Taxicogenomics Database
- RSEM:
-
RNA-seq by expectation maximization
- SD:
-
Standard deviation
- mRNA:
-
Messenger ribonucleic acid
- DNA:
-
Deoxyribonucleic acid
- GO:
-
Gene ontology
- GSEA:
-
Gene set enrichment analysis
- NES:
-
Normalised enrichment score
- FDR:
-
False discovery rate
- KEGG:
-
Key Kyoto Encyclopedia of Genes and Genomes
- TF:
-
Transcription factor
- miRNAs:
-
Micro ribonucleic acid
- GRN:
-
Gene-regulatory network
- TNF:
-
Tumour necrosis factor
- NF-κB:
-
Nuclear factor kappa B
- PPI:
-
Protein-protein interaction
- TFRC:
-
Transferrin receptor
- KDM6B:
-
Lysine demethylase 6B
- TSG:
-
Tumour suppressor gene
- TILs:
-
Tumour-infiltrating lymphocytes
- TGF:
-
Transforming growth factor
- TGFβ:
-
Transforming growth factor beta
- ATF4:
-
Activating transcription factor 4
- MBD2:
-
Methyl-CpG-binding domain protein 2
- PHD:
-
Prolyl hydroxylase
References
Padala, S. A., Barsouk, A., Thandra, K. C., Saginala, K., Mohammed, A., Vakiti, A., Rawla, P., & Barsouk, A. (2020). Epidemiology of renal cell carcinoma. World Journal of Oncology, 11(3), 79–87. https://doi.org/10.14740/wjon1279
Muglia, V. F., & Prando, A. (2015). Renal cell carcinoma: Histological classification and correlation with imaging findings. Radiologia Brasileira, 48(3), 166–174. https://doi.org/10.1590/0100-3984.2013.1927
Dudani, S., de Velasco, G., Wells, J. C., Gan, C. L., Donskov, F., Porta, C., Fraccon, A., Pasini, F., Lee, J. L., Hansen, A., Bjarnason, G. A., Beuselinck, B., Pal, S. K., Yuasa, T., Kroeger, N., Kanesvaran, R., Reaume, M. N., Canil, C., Choueiri, T. K., & Heng, D. Y. C. (2021). Evaluation of clear cell, papillary, and chromophobe renal cell carcinoma metastasis sites and association with survival. JAMA Network Open, 4(1), e2021869. https://doi.org/10.1001/jamanetworkopen.2020.21869
Shuch, B., Amin, A., Armstrong, A. J., Eble, J. N., Ficarra, V., Lopez-Beltran, A., Martignoni, G., Rini, B. I., & Kutikov, A. (2015). Understanding pathologic variants of renal cell carcinoma: Distilling therapeutic opportunities from biologic complexity. European Urology, 67(1), 85–97. https://doi.org/10.1016/j.eururo.2014.04.029
Gordan, J. D., Thompson, C. B., & Simon, M. C. (2007). HIF and C-Myc: Sibling rivals for control of cancer cell metabolism and proliferation. Cancer Cell, 12(2), 108–113. https://doi.org/10.1016/j.ccr.2007.07.006
Tang, S.-W., Chang, W.-H., Su, Y.-C., Chen, Y.-C., Lai, Y.-H., Wu, P.-T., Hsu, C.-I., Lin, W.-C., Lai, M.-K., & Lin, J.-Y. (2009). MYC Pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Letters, 273(1), 35–43. https://doi.org/10.1016/j.canlet.2008.07.038
Kaplan, E. L., & Meier, P. (1958). Nonparametric estimation from incomplete observations. Journal of American Statistical Association, 53(282), 457. https://doi.org/10.2307/2281868
Yu, G., Li, F., Qin, Y., Bo, X., Wu, Y., & Wang, S. (2010). GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics, 26(7), 976–978. https://doi.org/10.1093/bioinformatics/btq064
Subramanian, A., Tamayo, P., Mootha, V. K., Mukherjee, S., Ebert, B. L., Gillette, M. A., Paulovich, A., Pomeroy, S. L., Golub, T. R., Lander, E. S., & Mesirov, J. P. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, 102(43), 15545–15550. https://doi.org/10.1073/pnas.0506580102
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. https://doi.org/10.1101/gr.1239303
Zhou, Y., Zhou, B., Pache, L., Chang, M., Khodabakhshi, A. H., Tanaseichuk, O., Benner, C., & Chanda, S. K. (2019). Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nature Communications, 10(1), 1523. https://doi.org/10.1038/s41467-019-09234-6
Zhou, G., Soufan, O., Ewald, J., Hancock, R. E. W., Basu, N., & Xia, J. (2019). NetworkAnalyst 3.0: A visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Research, 47(W1), W234–W241. https://doi.org/10.1093/nar/gkz240
Liu, Z.-P., Wu, C., Miao, H., & Wu, H. (2015). RegNetwork: An integrated database of transcriptional and post-transcriptional regulatory networks in human and mouse. Database, 2015, bav095. https://doi.org/10.1093/database/bav095
Mattingly, C. J., Rosenstein, M. C., Colby, G. T., Forrest, J. N., Jr., & Boyer, J. L. (2006). The Comparative Toxicogenomics Database (CTD): A resource for comparative toxicological studies. Journal of Experimental Zoology Part A: Comparative Experimental Biology, 305A(9), 689–692. https://doi.org/10.1002/jez.a.307
Dan, H., Zhang, S., Zhou, Y., & Guan, Q. (2019). <p>DNA methyltransferase inhibitors: Catalysts for antitumour immune responses</P>. OncoTargets and Therapy, 12, 10903–10916. https://doi.org/10.2147/OTT.S217767
Moore, L. D., Le, T., & Fan, G. (2013). DNA methylation and its basic function. Neuropsychopharmacology, 38(1), 23–38. https://doi.org/10.1038/npp.2012.112
Ehrlich, M. (2009). DNA Hypomethylation in cancer cells. Epigenomics, 1(2), 239–259. https://doi.org/10.2217/epi.09.33
Camuzi, D., de Amorim, Í., Ribeiro Pinto, L., Oliveira Trivilin, L., Mencalha, A., & Soares Lima, S. (2019). Regulation is in the air: The relationship between hypoxia and epigenetics in cancer. Cells, 8(4), 300. https://doi.org/10.3390/cells8040300
Holderfield, M. T., & Hughes, C. C. W. (2008). Crosstalk between vascular endothelial growth factor, notch, and transforming growth factor-β in vascular morphogenesis. Circulation Research, 102(6), 637–652. https://doi.org/10.1161/CIRCRESAHA.107.167171
Chaturvedi, M. M., Sung, B., Yadav, V. R., Kannappan, R., & Aggarwal, B. B. (2011). NF-ΚB addiction and its role in cancer: ‘One size does not fit all.’ Oncogene, 30(14), 1615–1630. https://doi.org/10.1038/onc.2010.566
Yang, C. J., Lee, J.-W., & Chung, J. W. (2015). Influences of diabetes on hearing recovery in noise-exposed mice. Journal of Audiology and Otology, 19(3), 138–143. https://doi.org/10.7874/jao.2015.19.3.138
Liberti, M. V., & Locasale, J. W. (2016). The Warburg effect: How does it benefit cancer cells? Trends in Biochemical Sciences, 41(3), 211–218. https://doi.org/10.1016/j.tibs.2015.12.001
Liberzon, A., Birger, C., Thorvaldsdóttir, H., Ghandi, M., Mesirov, J. P., & Tamayo, P. (2015). The molecular signatures database hallmark gene set collection. Cell Systems, 1(6), 417–425. https://doi.org/10.1016/j.cels.2015.12.004
Rao, V. S., Srinivas, K., Sujini, G. N., & Kumar, G. N. S. (2014). Protein-protein interaction detection: Methods and analysis. International Journal of Proteomics, 2014, 1–12. https://doi.org/10.1155/2014/147648
Scott, D. E., Bayly, A. R., Abell, C., & Skidmore, J. (2016). Small molecules, big targets: Drug discovery faces the protein–protein interaction challenge. Nature Reviews Drug Discovery, 15(8), 533–550. https://doi.org/10.1038/nrd.2016.29
Yang, O. C. Y., Maxwell, P. H., & Pollard, P. J. (2013). Renal cell carcinoma: Translational aspects of metabolism and therapeutic consequences. Kidney International, 84(4), 667–681. https://doi.org/10.1038/ki.2013.245
Yamada, Y., Hidaka, H., Seki, N., Yoshino, H., Yamasaki, T., Itesako, T., Nakagawa, M., & Enokida, H. (2013). Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-myc oncogene in renal cell carcinoma. Cancer Science, 104(3), 304–312. https://doi.org/10.1111/cas.12072
Sumithra, B., Saxena, U., & Das, A. B. (2019). A comprehensive study on genome-wide coexpression network of KHDRBS1/Sam68 reveals its cancer and patient-specific association. Scientific Reports, 9(1), 11083. https://doi.org/10.1038/s41598-019-47558-x
Yang, J.-Y., & Hung, M.-C. (2009). A new fork for clinical application: Targeting forkhead transcription factors in cancer. Clinical Cancer Research, 15(3), 752–757. https://doi.org/10.1158/1078-0432.CCR-08-0124
Lee, T. I., & Young, R. A. (2013). Transcriptional regulation and its misregulation in disease. Cell, 152(6), 1237–1251. https://doi.org/10.1016/j.cell.2013.02.014
Iorio, M. V., & Croce, C. M. (2012). MicroRNA Dysregulation in cancer: Diagnostics, monitoring and therapeutics. A Comprehensive Review. EMBO Molecular Medicine, 4(3), 143–159. https://doi.org/10.1002/emmm.201100209
Wang, L.-H., Wu, C.-F., Rajasekaran, N., & Shin, Y. K. (2018). Loss of tumor suppressor gene function in human cancer: An overview. Cellular Physiology and Biochemistry, 51(6), 2647–2693. https://doi.org/10.1159/000495956
Hakimi, A. A., Chen, Y.-B., Wren, J., Gonen, M., Abdel-Wahab, O., Heguy, A., Liu, H., Takeda, S., Tickoo, S. K., Reuter, V. E., Voss, M. H., Motzer, R. J., Coleman, J. A., Cheng, E. H., Russo, P., & Hsieh, J. J. (2013). Clinical and pathologic impact of select chromatin-modulating tumor suppressors in clear cell renal cell carcinoma. European Urology, 63(5), 848–854. https://doi.org/10.1016/j.eururo.2012.09.005
Noon, A. P., Vlatković, N., Polański, R., Maguire, M., Shawki, H., Parsons, K., & Boyd, M. T. (2010). P53 and MDM2 in renal cell carcinoma. Cancer, 116(4), 780–790. https://doi.org/10.1002/cncr.24841
Hakimi, A. A., Reznik, E., Lee, C.-H., Creighton, C. J., Brannon, A. R., Luna, A., Aksoy, B. A., Liu, E. M., Shen, R., Lee, W., Chen, Y., Stirdivant, S. M., Russo, P., Chen, Y.-B., Tickoo, S. K., Reuter, V. E., Cheng, E. H., Sander, C., & Hsieh, J. J. (2016). An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell, 29(1), 104–116. https://doi.org/10.1016/j.ccell.2015.12.004
Liu, L., Guo, R., Zhang, X., Liang, Y., Kong, F., Wang, J., & Xu, Z. (2017). Loss of SETD2, but not H3K36me3, correlates with aggressive clinicopathological features of clear cell renal cell carcinoma patients. BioScience Trends, 11(2), 214–220. https://doi.org/10.5582/bst.2016.01228
Gordan, J. D., Lal, P., Dondeti, V. R., Letrero, R., Parekh, K. N., Oquendo, C. E., Greenberg, R. A., Flaherty, K. T., Rathmell, W. K., Keith, B., Simon, M. C., & Nathanson, K. L. (2008). HIF-α Effects on c-Myc distinguish two subtypes of sporadic VHL-deficient clear cell renal carcinoma. Cancer Cell, 14(6), 435–446. https://doi.org/10.1016/j.ccr.2008.10.016
Creighton, C. J., Morgan, M., Gunaratne, PH, Wheeler, D. A., et. al, (2013). Comprehensive Molecular Characterization of Clear Cell Renal Cell Carcinoma. Nature, 499 (7456), 43–49.https://doi.org/10.1038/nature12222
Abousaway, O., Rakhshandehroo, T., van den Abbeele, A. D., Kircher, M. F., & Rashidian, M. (2021). Noninvasive imaging of cancer immunotherapy. Nanotheranostics, 5(1), 90–112. https://doi.org/10.7150/ntno.50860
Li, T., Fu, J., Zeng, Z., Cohen, D., Li, J., Chen, Q., Li, B., & Liu, X. S. (2020). TIMER20 for analysis of tumor-infiltrating immune cells. Nucleic Acids Research, 48(W1), W509–W514. https://doi.org/10.1093/nar/gkaa407
Jochems, C., & Schlom, J. (2011). Tumor-infiltrating immune cells and prognosis: The potential link between conventional cancer therapy and immunity. Experimental Biology and Medicine, 236(5), 567–579. https://doi.org/10.1258/ebm.2011.011007
Irizarry, R. A., Wang, C., Zhou, Y., & Speed, T. P. (2009). Gene set enrichment analysis made simple. Statistical Methods in Medical Research, 18(6), 565–575. https://doi.org/10.1177/0962280209351908
Qin, S., Shi, X., Wang, C., Jin, P., & Ma, F. (2019). Transcription factor and MiRNA interplays can manifest the survival of CcRCC patients. Cancers (Basel), 11(11), 1668. https://doi.org/10.3390/cancers11111668
Vishnoi, K., Viswakarma, N., Rana, A., & Rana, B. (2020). Transcription factors in cancer development and therapy. Cancers (Basel), 12(8), 2296. https://doi.org/10.3390/cancers12082296
Antonangeli, F., Natalini, A., Garassino, M. C., Sica, A., Santoni, A., di Rosa, F. (2020). Regulation of PD- L1 Expression by NF-ΚB in Cancer. Frontiers in Immunology, 11. https://doi.org/10.3389/fimmu.2020.584626
Deleuze, A., Saout, J., Dugay, F., Peyronnet, B., Mathieu, R., Verhoest, G., Bensalah, K., Crouzet, L., Laguerre, B., Belaud-Rotureau, M.-A., Rioux-Leclercq, N., & Kammerer-Jacquet, S.-F. (2020). Immunotherapy in renal cell carcinoma: The future is now. International Journal of Molecular Sciences, 21(7), 2532. https://doi.org/10.3390/ijms21072532
Tran Janco, J. M., Lamichhane, P., Karyampudi, L., & Knutson, K. L. (2015). Tumor-infiltrating dendritic cells in cancer pathogenesis. The Journal of Immunology, 194(7), 2985–2991. https://doi.org/10.4049/jimmunol.1403134
Liu, Y., Hu, X., Han, C., Wang, L., Zhang, X., He, X., & Lu, X. (2015). Targeting tumor suppressor genes for cancer therapy. BioEssays, 37(12), 1277–1286. https://doi.org/10.1002/bies.201500093
Koshiji, M., Kageyama, Y., Pete, E. A., Horikawa, I., Barrett, J. C., & Huang, L. E. (2004). HIF-1α Induces cell cycle arrest by functionally counteracting Myc. The EMBO Journal, 23(9), 1949–1956. https://doi.org/10.1038/sj.emboj.7600196
Li, S., Wen, L., Hu, X., Wei, Q., & Dong, Z. (2021). HIF in Nephrotoxicity during cisplatin chemotherapy: Regulation, function and therapeutic potential. Cancers (Basel), 13(2), 180. https://doi.org/10.3390/cancers13020180
Acknowledgements
We thank Dr. Asim Bikas Das (associate professor; National Institute of Technology, Warangal, Telangana) for critically reading the manuscript and for his constructive comments. We acknowledge financial assistance from the National Institute of Technology, Warangal, for providing the computational facility and fellowship for Ph.D. students.
Author information
Authors and Affiliations
Contributions
Jyotsna Priyam collected and analysed the data with the help of Urmila Saxena. Urmila Saxena conceived and designed the study. Writing, reviewing, editing, and original draft preparation was done by Jyotsna Priyam and Urmila Saxena. Visualization, investigation, and supervision were done by Urmila Saxena.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Highlights
1. Myc expression was high in tumour samples in comparison to normal in KIPAN [kidney chromophobe (KICH) + kidney renal clear cell carcinoma (KIRC) + kidney renal papillary cell carcinoma (KIRP)] and KIRC (kidney renal clear cell carcinoma).
2. Higher expressions of Myc were associated with poor overall survival in renal cancers (KIPAN and KIRC).
3. Coexpressed genes and directly linked proteins, transcription factors, and miRNAs with Myc in KIPAN and KIRC are analysed.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Priyam, J., Saxena, U. Computational Gene Expression and Network Analysis of Myc Reveal Insights into Its Diagnostic and Prognostic Role in Subtypes of Renal Cancer. Appl Biochem Biotechnol 195, 4251–4276 (2023). https://doi.org/10.1007/s12010-023-04357-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04357-5