Abstract
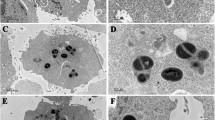
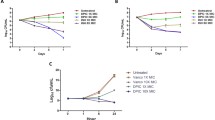
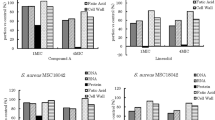
The incidences of methicillin-resistant strains of Staphylococcus aureus (MRSA) and their survival inside the macrophages are the major attributes of the relapsed infections after antimicrobial therapy, and it is a global problem. In this context, we have previously demonstrated 4-methoxy-1-methyl-2-oxopyridine-3-carbamide (MMOXC), a Ricinine derivative exhibiting anti-S. aureus and anti-biofilm characteristics by competitively inhibiting uridine monophosphate kinase (UMPK), UDP-N-acetyl muramyl pentapeptide ligase (Mur-F), and peptidyl deformylase, (PDF). In the present study, the stability of this competitive inhibitor MMOXC was evaluated by showing its ability to remain bound to the active sites of UMPK, Mur-F, and PDF even after increasing the incubation time, temperature, pH, and substrate concentration. On growing MRSA in fewer concentrations of MMOXC, these strains could not attain resistance to MMOXC and at the same time distinct reductions in the expression of UMPK, Mur-F, and PDF genes were noted. In vitro, infective models were generated by infecting MRSA to RAW 264.7 and human monocyte-derived macrophage (hMDM) cell lines. In these infected cell lines, in spite of increased nitric oxide synthase (NOS), NADPH–P450 reductase, superoxide dismutase, catalase, and peroxidase activities, the MRSA survived. At 640 µM/ml, the concentration of MMOXC penetrated into these infected cells and obliterated MRSA. While treating uninfected macrophage cell lines with MMOXC, no appreciable effect was observed indicating that MMOXC is the most suitable drug for the treatment of infections caused by MRSA.






Similar content being viewed by others
Data Availability
The data presented in the manuscript are available from the corresponding author who can be contacted at a reasonable time. Some of the data which are cloning of UMPK, Mur-f, and PDF are available at GenBank (accession no: FJ415072, JX841309, and JX311310).
Abbreviations
- ANAE :
-
Alpha-naphthyl acetate esterase
- ATCC :
-
American Type Culture Collection
- BHI :
-
Brain Heart Infusion
- BP medium :
-
Baird Parker Medium
- CA-MRSA :
-
Community-associated methicillin-resistant S. aureus
- CAT :
-
Catalase
- CFU :
-
Colony-forming unit
- d -ala- d -ala :
-
D-alanine-d-alanine
- DMEM :
-
Dulbecco’s modified Eagle medium
- IPTG :
-
Isopropyl-β-d-thiogalactopyranoside
- hMDMs :
-
Human monocyte-derived macrophages
- I :
-
Inhibitor concentration
- K i :
-
Inhibitor concentration required to achieve half maximum inhibition
- Km :
-
Michaelis Menten constant
- Km apparent :
-
Km of the reaction in the presence of an inhibitor
- Mϕ :
-
Macrophages
- MDR :
-
Multidrug-resistant
- MH agar :
-
Mueller Hinton Agar
- MMOXC :
-
4-Methoxy,1-methyl-2-oxo-pyridine,3-carbomide
- MOI :
-
Multiplicities of infection
- MRSA :
-
Methicillin resistant Staphylococcus aureus
- MSCRAMMs :
-
Microbial surface components recognizing adhesive matrix molecules
- Mur-F :
-
UDP-N-acetyl muramyl pentapeptide ligase
- MTT :
-
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- NOS :
-
Nitric oxide synthase
- LB media :
-
Luria Bertini media
- PDF :
-
Peptidyl deformylase
- RAW 264.7 :
-
Ralph and William’s cell line 264.7
- RNS :
-
Reactive nitrogen species
- ROS :
-
Reactive oxygen species
- SOD :
-
Superoxide dismutase
- THP-1 cell line :
-
Human monocytic cell line
- UMPK :
-
Uridine monophosphate kinase
- UMT :
-
UDP-N-acetyl muramic acid tripeptide
- PBS :
-
Phosphate-buffered saline
- PCR :
-
Polymerase chain reaction
- PMA :
-
Phorbol 12-myristate 13-acetate
- RPMI :
-
Roswell Park Memorial Institute
References
Thomer, L., Schneewind, O., & Missiakas, D. (2016). Pathogenesis of Staphylococcus aureus bloodstream infections. Annual Review of Pathology: Mechanisms of Disease, 11, 343–364.
Laupland, K. B., & Church, D. L. (2014). Population-based epidemiology and microbiology of community-onset bloodstream infections. Clinical Microbiology Reviews, 27(4), 647–664.
DeLeo, F. R., Otto, M., Kreiswirth, B. N., & Chambers, H. F. (2010). Community-associated meticillin-resistant Staphylococcus aureus. Lancet, 375(9725), 1557–1568.
Turner, N. A., Sharma-Kuinkel, B. K., Maskarinec, S. A., Eichenberger, E. M., Shah, P. P., Carugati, M., Holland, T. L., & Fowler, V. G. (2019). Methicillin-resistant Staphylococcus aureus: An overview of basic and clinical research. Nature Reviews Microbiology, 17(4), 203–218.
Somerville, G. A., & Proctor, R. A. (2009). At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiology and Molecular Biology Reviews, 73(2), 233–248.
Tuchscherr, L., Löffler, B., & Proctor, R. A. (2020). Persistence of Staphylococcus aureus: Multiple metabolic pathways impact the expression of virulence factors in small-colony variants (SCVs). Frontiers in Microbiology, 11, 1028.
Rudra, P., & Boyd, J. M. (2020). Metabolic control of virulence factor production in Staphylococcus aureus. Current Opinion in Microbiology, 55, 81–87.
Foster, T. J., Geoghegan, J. A., Ganesh, V. K., & Höök, M. (2014). Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nature Reviews Microbiology, 12(1), 49–62.
Sendi, P., & Proctor, R. A. (2009). Staphylococcus aureus as an intracellular pathogen: The role of small colony variants. Trends in Microbiology, 17(2), 54–58.
Proctor, R. A., Kriegeskorte, A., Kahl, B. C., Becker, K., Löffler, B., & Peters, G. (2014). Staphylococcus aureus Small Colony Variants (SCVs): A road map for the metabolic pathways involved in persistent infections. Frontiers in Cellular and Infection Microbiology, 4, 99.
Thammavongsa, V., Kim, H. K., Missiakas, D., & Schneewind, O. (2015). Staphylococcal manipulation of host immune responses. Nature Reviews Microbiology, 13(9), 529–543.
Torlak, E., Korkut, E., Uncu, A. T., & Şener, Y. (2017). Biofilm formation by Staphylococcus aureus isolates from a dental clinic in Konya, Turkey. Journal of Infection and Public Health, 10(6), 809–813.
Amaral, M. M., Coelho, L. R., Flores, R. P., Souza, R. R., Silva-Carvalho, M. C., Teixeira, L. A., Ferreira-Carvalho, B. T., & Figueiredo, A. M. (2005). The predominant variant of the Brazilian epidemic clonal complex of methicillin-resistant Staphylococcus aureus has an enhanced ability to produce biofilm and to adhere to and invade airway epithelial cells. Journal of Infectious Diseases, 192(5), 801–810.
Yeswanth, S., Chaudhury, A., & Sarma, P. (2017). Quantitative expression analysis of SpA, FnbA and Rsp genes in Staphylococcus aureus: Actively associated in the formation of biofilms. Current Microbiology, 74(12), 1394–1403.
Swarupa, V., Chaudhury, A., & Sarma, P. (2018). Iron enhances the peptidyl deformylase activity and biofilm formation in Staphylococcus aureus. 3 Biotech, 8(1), 32.
Vudhya Gowrisankar, Y., Manne Mudhu, S., Pasupuleti, S. K., Suthi, S., Chaudhury, A., & Sarma, P. (2021). Staphylococcus aureus grown in anaerobic conditions exhibits elevated glutamine biosynthesis and biofilm units. Canadian Journal of Microbiology, 67(4), 323–331.
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P., & Hall-Stoodley, L. (2017). Targeting microbial biofilms: current and prospective therapeutic strategies. Nature Reviews Microbiology, 15, 740–755.
Silva, M. T. (2010). When two is better than one: Macrophages and neutrophils work in concert in innate immunity as complementary and cooperative partners of a myeloid phagocyte system. Journal of Leukocyte Biology, 87(1), 93–106.
Bogdan, C., Röllinghoff, M., & Diefenbach, A. (2000). Reactive oxygen and reactive nitrogen intermediates in innate and specific immunity. Current Opinion in Immunology, 12(1), 64–76.
Herb, M., & Schramm, M. (2021). Functions of ROS in macrophages and antimicrobial immunity. Antioxidants, 10(2), 313.
Hommes, J. W., & Surewaard, B. (2022). Intracellular habitation of Staphylococcus aureus: Molecular mechanisms and prospects for antimicrobial therapy. Biomedicines, 10(8), 1804.
Geoghegan, J. A., & Foster, T. J. (2017). Cell wall-anchored surface proteins of Staphylococcus aureus: Many proteins, multiple functions. Current Topics in Microbiology and Immunology, 409, 95–120.
Foster, T. J. (2019). The MSCRAMM family of cell-wall-anchored surface proteins of Gram-positive cocci. Trends in Microbiology, 27, 927–941.
Schilcher, K., & Horswill, A. R. (2020). Staphylococcal biofilm development: Structure, regulation, and treatment strategies. Microbiology and Molecular Biology Reviews, 84(3), e00026–19.
Antimicrobial Resistance Collaborators. (2022). Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet, 399(10325), 629–655.
Dey, J., Mahapatra, S. R., Raj, T. K., Kaur, T., Jain, P., Tiwari, A., Patro, S., Misra, N., & Suar, M. (2022). Designing a novel multi-epitope vaccine to evoke a robust immune response against pathogenic multidrug-resistant Enterococcus faecium bacterium. Gut Pathogens. 14(1), 21.
Dey, J., Mahapatra, S. R., Lata, S., Patro, S., Misra, N., & Suar, M. (2022). Exploring Klebsiella pneumoniae capsule polysaccharide proteins to design multiepitope subunit vaccine to fight against pneumonia. Expert Review of Vaccines, 21(4), 569–587.
Mahapatra, S. R., Dey, J., Kushwaha, G. S., Puhan, P., Mohakud, N. K., Panda, S. K., Lata, S., Misra, N., & Suar, M. (2021). Immunoinformatic approach employing modeling and simulation to design a novel vaccine construct targeting MDR efflux pumps to confer wide protection against typhoidal. Journal of Biomolecular Structure and Dynamics, 31, 1–13.
Dey, J., Mahapatra, S. R., Patnaik, S., Lata, S., Kushwaha, G. S., Panda, R. K., Misra, N., & Suar, M. (2022). Molecular characterization and designing of a novel multiepitope vaccine construct against Pseudomonas aeruginosa. International Journal of Peptide Research and Therapeutics, 28(2), 49.
Mahapatra, S. R., Dey, J., Raj, T. K., Kumar, V., Ghosh, M., Verma, K. K., Kaur, T., Kesawat, M. S., Misra, N., & Suar, M. (2022). The potential of plant-derived secondary metabolites as novel drug candidate against Klebsiella pneumoniae: molecular docking and simulation investigation. South African Journal of Botany, 149, 789–797.
Narang, P. K., Dey, J., Mahapatra, S. R., Ghosh, M., Misra, N., Suar, M., Kumar, V., & Raina, V. (2021). Functional annotation and sequence-structure characterization of a hypothetical protein putatively involved in carotenoid biosynthesis in microalgae. South African Journal of Botany, 141, 219–226.
Narang, P. K., Dey, J., Mahapatra, S. R., Roy, R., Kushwaha, G. S., Misra, N., Suar, M., & Raina, V. (2021). Genome-based identification and comparative analysis of enzymes for carotenoid biosynthesis in microalgae. World Journal of Microbiology & Biotechnology, 38(1), 8.
Arora, D. S., & Mahajan, H. (2019). Major phytoconstituents of Prunus cerasoides responsible for antimicrobial and antibiofilm potential against some reference strains of pathogenic bacteria and clinical isolates of MRSA. Applied Biochemistry and Biotechnology, 188(4), 1185–1204.
Sobral, R. G., Ludovice, A. M., de Lencastre, H., & Tomasz, A. (2006). Role of murF in cell wall biosynthesis: Isolation and characterization of a murF conditional mutant of Staphylococcus aureus. Journal of Bacteriology, 188(7), 2543–2553.
Hari Prasad, O., Nanda Kumar, Y., Reddy, O. V., Chaudhary, A., & Sarma, P. V. (2012). Cloning, expression, purification and characterization of UMP kinase from Staphylococcus aureus. The Protein Journal, 31(4), 345–352.
Mader, D., Liebeke, M., Winstel, V., Methling, K., Leibig, M., Götz, F., Lalk, M., & Peschel, A. (2013). Role of N-terminal protein formylation in central metabolic processes in Staphylococcus aureus. BMC Microbiology, 13, 7.
Swarupa, V., Chaudhury, A., & Krishna Sarma, P. V. (2017). Effect of 4-methoxy 1-methyl 2-oxopyridine 3-carbamide on Staphylococcus aureus by inhibiting UDP-MurNAc-pentapeptide, peptidyl deformylase and uridine monophosphate kinase. Journal of Applied Microbiology, 122(3), 663–675.
El-Naggar, M. H., Elgaml, A., Abdel Bar, F. M., & Badria, F. A. (2019). Antimicrobial and antiquorum-sensing activity of Ricinus communis extracts and ricinine derivatives. Natural Product Research, 33(11), 1556–1562.
Janská, P., Knejzlík, Z., Perumal, A. S., Jurok, R., Tokárová, V., Nicolau, D. V., Štěpánek, F., & Kašpar, O. (2021). Effect of physicochemical parameters on the stability and activity of garlic alliinase and its use for in-situ allicin synthesis. PloS one, 16(3), e0248878.
El-Shaheny, R. N., & Yamada, K. (2015). The influence of pH and temperature on the stability of flutamide. An HPLC investigation and identification of the degradation product by EI+-MS. RSC Advances, 5(5), 3206–3214.
Bailey, J., Douglas, H., de Masino, L., Carvalho, L., & Argyrou, A. (2021). Human Mat2A uses an ordered kinetic mechanism and is stabilized but not regulated by Mat2B. Biochemistry, 60(47), 3621–3632.
Fraunholz, M., & Sinha, B. (2012). Intracellular Staphylococcus aureus: Live-in and let die. Frontiers in Cellular and Infection Microbiology, 2, 43.
Strobel, M., Pförtner, H., Tuchscherr, L., Völker, U., Schmidt, F., Kramko, N., Schnittler, H. J., Fraunholz, M. J., Löffler, B., Peters, G., & Niemann, S. (2016). Post-invasion events after infection with Staphylococcus aureus are strongly dependent on both the host cell type and the infecting S. aureus strain. Clinical Microbiology and Infection, 22(9), 799–809.
Tuchscherr, L., Heitmann, V., Hussain, M., Viemann, D., von Roth, J., Eiff, C., Peters, G., Becker, K., & Löffler, B. (2010). Staphylococcus aureus small-colony variants are adapted phenotypes for intracellular persistence. Journal of Infectious Diseases, 202(7), 1031–1040.
Tuchscherr, L., Medina, E., Hussain, M., Völker, W., Heitmann, V., Niemann, S., Holzinger, D., Roth, J., Proctor, R. A., Becker, K., Peters, G., & Löffler, B. (2011). Staphylococcus aureus phenotype switching: An effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Molecular Medicine, 3(3), 129–141.
Lu, H., & Tonge, P. J. (2010). Drug-target residence time: Critical information for lead optimization. Current Opinion in Chemical Biology, 14, 467–474.
Koziel, J., Maciag-Gudowska, A., Mikolajczyk, T., Bzowska, M., Sturdevant, D. E., Whitney, A. R., Shaw, L. N., DeLeo, F. R., & Potempa, J. (2009). Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS One, 4(4), e5210.
Lacoma, A., Cano, V., Moranta, D., Regueiro, V., Domínguez-Villanueva, D., Laabei, M., González-Nicolau, M., Ausina, V., Prat, C., & Bengoechea, J. A. (2017). Investigating intracellular persistence of Staphylococcus aureus within a murine alveolar macrophage cell line. Virulence, 8(8), 1761–1775.
Starr, T., Bauler, T. J., Malik-Kale, P., & Steele-Mortimer, O. (2018). The phorbol 12-myristate-13-acetate differentiation protocol is critical to the interaction of THP-1 macrophages with Salmonella Typhimurium. PloS One, 13(3), e0193601.
Tedesco, S., De-Majo, F., Kim, J., Trenti, A., Trevisi, L., Fadini, G. P., Bolego, C., Zandstra, P. W., Cignarella, A., & Vitiello, L. (2018). Convenience versus biological significance: are PMA-differentiated THP-1 cells a reliable substitute for blood-derived macrophages when studying in Vitro polarization? Frontiers in Pharmacology, 9, 71.
Carrasco-Pozo, C., Tan, K. N., & Avery, V. M. (2020). Hemin prevents increased glycolysis in macrophages upon activation: protection by microbiota-derived metabolites of polyphenols. Antioxidants, 9(11), 1109.
Monahan, R. A., Dvorak, H. F., & Dvorak, A. M. (1981). Ultrastructural localization of nonspecific esterase activity in guinea pig and human monocytes, macrophages, and lymphocytes. Blood, 58(6), 1089–1099.
Amano, F., & Noda, T. (1995). Improved detection of nitric oxide radical (NO.) production in an activated macrophage culture with a radical scavenger, carboxy PTIO and Griess reagent. FEBS Letters, 368(3), 425–428.
Guengerich, F. P., Martin, M. V., Sohl, C. D., & Cheng, Q. (2009). Measurement of cytochrome P450 and NADPH-cytochrome P450 reductase. Nature Protocols, 4(9), 1245–1251.
Weydert, C. J., & Cullen, J. J. (2010). Measurement of superoxide dismutase, catalase and glutathione peroxidase in cultured cells and tissue. Nature Protocols, 5(1), 51–66.
van Meerloo, J., Kaspers, G. J., & Cloos, J. (2011). Cell sensitivity assays: The MTT assay. Methods in Molecular Biology, 731, 237–245.
Weaver, A. J., Peters, T. R., Tripet, B., Van Vuren, A., Rakesh Lee, R. E., Copié, V., & Teintze, M. (2018). Exposure of methicillin-resistant Staphylococcus aureus to low levels of the antibacterial THAM-3ΦG generates a small colony drug-resistant phenotype. Scientific Reports, 8(1), 9850.
Dey, S., & Bishayi, B. (2015). Killing of Staphylococcus aureus in murine macrophages by chloroquine used alone and in combination with ciprofloxacin or azithromycin. Journal of Inflammation Research, 8, 29–47.
Pidwill, G. R., Gibson, J. F., Cole, J., Renshaw, S. A., & Foster, S. J. (2021). The role of macrophages in Staphylococcus aureus infection. Frontiers in Immunology, 11, 620339.
Acknowledgements
We sincerely acknowledge the Sri Venkateswara Institute of Medical Sciences and University for providing facilities to carry out this work. This work forms part of the PhD work to be submitted to SVIMS University.
Funding
This work was supported by a Sri Balaji Arogya Vara Prasadini (SBAVP) scheme (Grant No. SBAVP/PhD/28 dated: 30th December 2019).
Author information
Authors and Affiliations
Contributions
PVGK conceived the idea and design of the experiments; SSR was involved in the development and optimization of experimental methods; DPK participated in the microbial experiments; SSR and PVGK participated in the analysis and interpretation of data; PVGK, SSR, and ACH worked together to prepare the manuscript and revised the manuscript.
Corresponding author
Ethics declarations
Ethical Approval
This study was cleared by the institutional ethics committee (IEC. No: 960, dated: 18 September, 2019).
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Suthi, S., Gopi, D., Chaudhary, A. et al. The Therapeutic Potential of 4-Methoxy-1-methyl-2-oxopyridine-3-carbamide (MMOXC) Derived from Ricinine on Macrophage Cell Lines Infected with Methicillin-Resistant Strains of Staphylococcus aureus. Appl Biochem Biotechnol 195, 2843–2862 (2023). https://doi.org/10.1007/s12010-022-04269-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04269-w