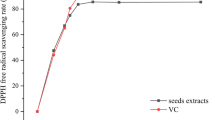
Abstract
Breast cancer is one of the most commonly diagnosed cancers in woman which accounts for more than 1 in 10 new cancers in the entire world. The recently found four new potential hub genes that show a strong expression in breast cancer are CCNA2, CCNB1, MAD2L1, and RAD51. Nowadays, food habits and lifestyle of an individual are one of the factors for causing cancers. Consumption of seeds on a regular basis is the key factor for leading a good health. Sesame seeds and Sunflower seeds are few examples of cancer fighting seeds. Sesame (Sesamum indicum) is one of the earliest oil seed plant with various phytocompounds present which include lignans, tocopherols, phenolics, polyunsaturated fatty acids, and phytosterols. Sunflower (Helianthus annuus L.) is primarily harvested as an oil seed plant with various phytocompounds present which include flavonoids, phenolic acids, tocopherols, and vitamin B3. These are the few seeds that help women to prevent and also to fight against Breast cancer with its potential anti-cancer activity. The main objective of the current study is to identify the potential phytocompounds present in the cancer fighting seeds using molecular docking and dynamic simulation approach which can further help pharmaceuticals industries in producing targeted drugs against breast cancer hub genes as well as food industries in producing products combining the potential phytocompounds present in the seeds.




















Similar content being viewed by others
Data availability
Data will be available on request.
Code availability
Not applicable.
References
Ong, C. H. C., Lee, D. Y., Lee, B., et al. (2022). Hypoxia-regulated carbonic anhydrase IX (CAIX) protein is an independent prognostic indicator in triple negative breast cancer. Breast Cancer Research, 24, 38. https://doi.org/10.1186/s13058-022-01532-0
Minoura, Y., Takahashi, M., Maeda, H., et al. (2022). Significance of prostate/pancreatic/skin cancer family history for detecting BRCA2 pathogenic variant careers among patients with breast cancer. Breast Cancer. https://doi.org/10.1007/s12282-022-01360-2
Raghavakaimal, A., Cristofanilli, M., Tang, C. M., et al. (2022). CCR5 activation and endocytosis in circulating tumor-derived cells isolated from the blood of breast cancer patients provide information about clinical outcome. Breast Cancer Research, 24, 35. https://doi.org/10.1186/s13058-022-01528-w
Dashti, S., Taheri, M., & Ghafouri-Fard, S. (2020). An in-silico method leads to recognition of hub genes and crucial pathways in survival of patients with breast cancer. Science and Reports, 10, 18770. https://doi.org/10.1038/s41598-020-76024-2
Deng, J.-L., Yun-hua, Xu., & Wang, G. (2019). Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Frontiers in Genetics. https://doi.org/10.3389/fgene.2019.00695
Gao, Tian & Han, Yong & Yu, Ling & Ao, Sheng & li, ZiYu & ji, Jiafu. CCNA2 is a prognostic biomarker for ER+ breast cancer and tamoxifen resistance. PloS one. 9. e9177 (2014). https://doi.org/10.1371/journal.pone.0091771
Wang, Y., Zhong, Q., Zhaoyun, Li., Lin, Z., Chen, H., & Wang, P. (2021). Integrated profiling identifies CCNA2 as a potential biomarker of immunotherapy in breast cancer. OncoTargets and Therapy., 14, 2433–2448. https://doi.org/10.2147/ott.s296373
Ding, Kun & Li, Wenqing & Zou, Zhiqiang & Zou, Xianzhi & Wang, Chengru. CCNB1 is a prognostic biomarker for ER+ breast cancer. Medical hypotheses. 83. (2014). https://doi.org/10.1016/j.mehy.2014.06.013
Fang, L.; Liu, Q.; Cui, H.; Zheng, Y.; Wu, C. Bioinformatics analysis highlight differentially expressed CCNB1 and PLK1 genes as potential anti-breast cancer drug targets and prognostic markers. Genes,13,654. (2022) https://doi.org/10.3390/genes13040654
Wang, Zhanwei & Katsaros, Dionyssios & Shen, yi & Fu, Yuanyuan & Canuto, Emilie & Benedetto, Chiara & Lu, Lingeng & Chu, Wen-Ming & Risch, Harvey & Yu, Herbert. Biological and clinical significance of MAD2L1 and BUB1, genes frequently appearing in expression signatures for breast cancer prognosis. PloS one. 10. e0136246. (2015). https://doi.org/10.1371/journal.pone.0136246
Tsantoulis, Petros & Migliorini, Denis & Martin Lluesma, Silvia & Durigova, Anna & Bodmer, A. & Dietrich, P. & Labidi-Galy, Intidhar. Mad2L1 overexpression leads to early metastasis in breast cancer. Annals of oncology : official journal of the European Society for Medical Oncology. (2014). https://doi.org/10.1093/annonc/mdu327.29
Wu, R., Patel, A., Tokumaru, Y., et al. (2022). High RAD51 gene expression is associated with aggressive biology and with poor survival in breast cancer. Breast Cancer Research and Treatment, 193, 49–63. https://doi.org/10.1007/s10549-022-06552-0
Wiegmans, Adrian & Al-Ejeh, Fares & Chee, Nicole & Yap, Pei-Yi & Gorski, Julia & Da Silva, Leonard & Bolderson, Emma & Chenevix-Trench, Georgia & Anderson, Robin & Simpson, Peter & Lakhani, Sr & Khanna, Kum. Rad51 supports triple negative breast cancer metastasis. Oncotarget. 5. 10. (2014). https://doi.org/10.18632/oncotarget.1923
Pathak N., Bhaduri A., Rai A.K. Sesame: bioactive compounds and health benefits. In: Mérillon JM., Ramawat K. (eds) Bioactive Molecules in Food. Reference Series in Phytochemistry. Springer, Cham. (2019) https://doi.org/10.1007/978-3-319-78030-6_59
Lin Zhou, Xiaohui Lin, Arshad Mehmood Abbasi, Bisheng Zheng, "Phytochemical contents and antioxidant and antiproliferative activities of selected black and white sesame seeds", BioMed Research International, vol. 2016, Article ID 8495630, 9 pages, 2016. https://doi.org/10.1155/2016/8495630
Guo S, Ge Y, Na Jom K. A review of phytochemistry, metabolite changes, and medicinal uses of the common sunflower seed and sprouts (Helianthus annuus L.). Chem Cent J. 2017;11(1):95. Published 2017 https://doi.org/10.1186/s13065-017-0328-7
Anitha, S., Ranjani, S., & Hemalatha, S. (2021). In silico analysis of quercetin and its analogues against targeted Proteins. Bio interface Research in Applied Chemistry, 11(5), 13695–13705.
Abinaya, M., Priya, S., Karunya, J. R., Ranjani, S., & Hemalatha, S. (2021). Screening the efficacy of compounds from ghee to control cancer: An in silico approach. Biointerface Research in Applied Chemistry, 11(6), 14115–14126. https://doi.org/10.33263/BRIAC116.1411514126
Venkatesan, H., Soundhararajan, R., & Srinivasan, H. (2022). Interaction of various types of bisphenols with enzymes involved in melanin synthesis. Toxicol Environ Health Sci, 14, 19–24. https://doi.org/10.1007/s13530-021-00111-8
Asma, B., & Hemalatha, S. (2022). SARS-CoV-2 inhibitors from Nigella sativa. Applied Biochemistry and Biotechnology, 194, 1051–1090. https://doi.org/10.1007/s12010-021-03790-8
Abraham JT, Maharifa HNS, Hemalatha S. In silico molecular docking approach against enzymes causing Alzheimer’s disease using Borassus flabellifer Linn. Appl Biochem Biotechnol. Published online 2022:1–10. https://doi.org/10.1007/s12010-021-03779-3
Mendie, L. E., & Hemalatha, S. (2022). Molecular docking of phytochemicals targeting GFRs as therapeutic sites for cancer: An in silico study. Applied Biochemistry and Biotechnology, 194(1), 215–231. https://doi.org/10.1007/s12010-021-03791-7
Dallakyan, S., & Olson, A. J. (2015). Small-molecule library screening by docking with PyRx. Methods Mol Biol, 1263, 243–50. https://doi.org/10.1007/978-1-4939-2269-7_19
Ravindranath, K.J., Mohaideen, N.S.M.H. & Srinivasan, H. Phytocompounds of onion target heat shock proteins (HSP70s) to control breast cancer malignancy. Appl Biochem Biotechnol(2022).https://doi.org/10.1007/s12010-022-04016-1
Souzia S Ranjani S , Hemalatha S Repurposing of drugs targeted against COVID-19 spike receptor for treatment: An in silico approach 2021, Bio interface Research in Applied Chemistry, 11, (5) https://doi.org/10.33263/BRIAC115.1374013753
Kuriata, A., Gierut, A. M., Oleniecki, T., Ciemny, M. P., Kolinski, A., Kurcinski, M., & Kmiecik, S. (2018). CABS-flex 20 A web server for fast simulations of flexibility of protein structures. Nucleic Acids Research, 46(1), 338–343.
Vardhini, S. P., Sadiya, H., Beig, S., Pandurangan, A. K., Hemelatha, S., & Waseem, M. (2021). Possible interaction of non-steroidal anti-inflammatory drugs against NF-κB and COX-2 mediated inflammation: An in silico probe. Applied Biochemistry and Biotechnology. https://doi.org/10.1007/s12010-021-03719-1
Arpudhamary, V., Priya, S., Muhammed, A. P., Manzoor, D., Mubarakali, S., & Hemalatha. (2019). Apoptotic-inducing factor 1 (AIF1) is critical in cembranoid mediated apoptosis to control cancer. Biocatalysis Agricultural biotechnology, 22, 101343. https://doi.org/10.1016/j.bcab.2019.101343
Acknowledgements
The authors are very much grateful to School of Life Sciences, B S Abdur Rahman Crescent Institute of Science and Technology, Chennai, for providing research facilities and also for their constant support and encouragement.
Author information
Authors and Affiliations
Contributions
SH conceived and designed research. SA and NS conducted experiments and analyzed data. All authors wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
All authors read and approved the manuscript for publication.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kumar, S.A., Mohaideen, N.S.M.H. & S, H. Phytocompounds From Edible Oil Seeds Target Hub Genes To Control Breast Cancer. Appl Biochem Biotechnol 195, 1231–1254 (2023). https://doi.org/10.1007/s12010-022-04224-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-04224-9