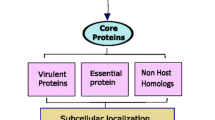
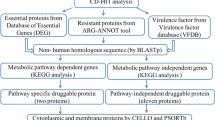
Abstract
Multidrug-resistant Acinetobacter baumannii (A. baumannii) infections are becoming more prevalent all over the world. As a cost-effective and preventative method, vaccination seems to be required against this bacterium. In the present study, subtractive proteomics along with reverse vaccinology approaches was used to predict suitable therapeutics against A. baumannii. Using the Vaxign online tool, we studied over 35 genomes of A. baumannii strains and chose outer membrane and secreted proteins of A. baumannii 1656–2 as possible vaccine candidates. Then, investigations were performed on the immunogenicity, antigenic characteristics, physicochemical properties, B-cell and MHC class I, and MHC class II molecules epitope densities of proteins. After optimizing the codon of the proteins, the pcDNA3.1( +) expression construct was designed and the immunogenicity, allergenicity, and physicochemical properties of the vaccine construct were predicted. Hcp and OmpC proteins were predicted as extracellular and outer membrane proteins, respectively. These proteins interact with 10 other proteins to form a network of protein interactions with virulence properties. Immunoassays of Hcp and OmpC proteins showed antigenicity of 0.88 and 0.79, respectively. These proteins have 5 structural cell epitope points and 5 linear B epitope points. They are also able to bind to different HLA alleles of MCH class I/class II as selected immunogenic proteins and designed non-allergenic structures with solubility of 0.650 and immunogenicity score of 0.91. The results of this “in silico” study indicate high specificity and the development of a significant humoral and cellular immune response. It can be concluded that the Hcp and OmpC dual vaccine construct is one of the promising candidates against A. baumannii. The findings of this “in silico” study show excellent specificity and the emergence of a substantial humoral and cellular immune response. This is a computer-based study that needs to be tested in vitro and in vivo to corroborate the conclusions of the vaccine design procedures.







Similar content being viewed by others
Data Availability
The datasets analyzed during the current study are available from the corresponding author on reasonable request.
References
Longo, F., Vuotto, C., & Donelli, G. (2014). Biofilm formation in Ab. New Microbiologica, 37(2), 119–127.
Wang, X., & Qin, L. J. (2019). A review on Ab. Journal of Acute Disease, 8(1), 16.
Morris, F. C., Dexter, C., Kostoulias, X., Uddin, M. I., & Peleg, A. Y. (2019). The mechanisms of disease caused by Ab. Frontiers in microbiology, 10, 1601.
Zeighami, H., Valadkhani, F., Shapouri, R., Samadi, E., & Haghi, F. (2019). Virulence characteristics of multidrug resistant biofilm forming Ab isolated from intensive care unit patients. BMC infectious diseases, 19(1), 1–9.
Gharaghie, T. P., Beiranvand, S., Riahi, A., Badmasti, F., Shirin, N. J., Mirzaie, A., Elahianfar, Y., Ghahari, S., Ghahari, S., Pasban, K., & Hajrasoliham, S. (2022). Fabrication and characterization of thymol‐loaded chitosan nanogels: Improved antibacterial and anti‐biofilm activities with negligible cytotoxicity. Chemistry & Biodiversity, 19(3), e202100426.
Samsudin, F., Ortiz-Suarez, M. L., Piggot, T. J., Bond, P. J., & Khalid, S. (2016). OmpA: A flexible clamp for bacterial cell wall attachment. Structure, 24(12), 2227–2235.
Ferenci, T., & Phan, K. (2015). How porin heterogeneity and trade-offs affect the antibiotic susceptibility of Gram-negative bacteria. Genes, 6(4), 1113–1124.
Galdiero, S., Falanga, A., Cantisani, M., Tarallo, R., Della Pepa, M. E., D’Oriano, V., & Galdiero, M. (2012). Microbe-host interactions: structure and role of Gram-negative bacterial porins. Current Protein and Peptide Science, 13(8), 843–854.
Darcan, C. (2012). Expression of OmpC and OmpF porin proteins and survival of Escherichia coli under photooxidative stress in Black Sea water. Aquatic Biology, 17(2), 97–105.
Arockiasamy, A., & Krishnaswamy, S. (2000). Purification of integral outer-membrane protein OmpC, a surface antigen from Salmonella typhi for structure–function studies: A method applicable to enterobacterial major outer-membrane protein. Analytical Biochemistry, 283(1), 64–70.
Zawawi, A., Forman, R., Smith, H., Mair, I., Jibril, M., Albaqshi, M. H., Brass, A., Derrick, J. P., & Else, K. J. (2020). In-Silico design of a T-cell epitope vaccine candidate for parasitic helminth infection. PLoS pathogens, 16(3), e1008243.
Pérez-Toledo, M., Valero-Pacheco, N., Pastelin-Palacios, R., Gil-Cruz, C., Perez-Shibayama, C., Moreno-Eutimio, M. A., Becker, I., Pérez-Tapia, S. M., Arriaga-Pizano, L., Cunningham, A. F., & Isibasi, A. (2017). Salmonella Typhi porins OmpC and OmpF are potent adjuvants for T-dependent and T-independent antigens. Frontiers in immunology, 8, 230.
Zoued, A., Brunet, Y. R., Durand, E., Aschtgen, M. S., Logger, L., Douzi, B., Journet, L., Cambillau, C., & Cascales, E. (2014). Architecture and assembly of the Type VI secretion system. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1843(8), 1664–1673.
Cascales, E., & Cambillau, C. (2012). Structural biology of type VI secretion systems. Philosophical Transactions of the Royal Society B: Biological Sciences, 367(1592), 1102–1111.
Laxminarayan, R., Duse, A., Wattal, C., Zaidi, A. K., Wertheim, H. F., Sumpradit, N., Vlieghe, E., Hara, G. L., Gould, I. M., Goossens, H., & Greko, C. (2013). Antibiotic resistance—The need for global solutions. The Lancet infectious diseases, 13(12), 1057–1098.
Afrough, B., Dowall, S., & Hewson, R. (2019). Emerging viruses and current strategies for vaccine intervention. Clinical & Experimental Immunology, 196(2), 157–166.
Bazmara, H., Rasooli, I., Jahangiri, A., Sefid, F., Astaneh, S. D. A., & Payandeh, Z. (2019). Antigenic properties of iron regulated proteins in Ab: An in-silico approach. International Journal of Peptide Research and Therapeutics, 25(1), 205–213.
Leow, C. Y., Kazi, A., Ismail, C. M. K. H., Chuah, C., Lim, B. H., Leow, C. H., & Singh, K. K. B. (2020). Reverse vaccinology approach for the identification and characterization of outer membrane proteins of Shigella flexneri as potential cellular-and antibody-dependent vaccine candidates. Clinical and experimental vaccine research, 9(1), 15–25.
Nezafat, N., Eslami, M., Negahdaripour, M., Rahbar, M. R., & Ghasemi, Y. (2017). Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Molecular BioSystems, 13(4), 699–713.
Habibi, N., Hashim, S. Z. M., Norouzi, A., & Samian, M. R. (2014). A review of machine learning methods to predict the solubility of overexpressed recombinant proteins in Escherichia coli. BMC Bioinformatics, 15(1), 1–16.
Sadat, S. A. (2018). The inhibitory effects of silver nanoparticles on bap gene expression in antibiotic-resistant Acientobacter bumanni isolates using real-time PCR. Journal of Ilam University of Medical Sciences, 26(4), 175–185.
Gul, S., Ahmad, S., Ullah, A., Ismail, S., Khurram, M., Tahir ul Qamar, M., Hakami, A. R., Alkhathami, A. G., Alrumaihi, F., & Allemailem, K. S. (2022). Designing a recombinant vaccine against Providencia rettgeri using immunoinformatics approach. Vaccines, 10, 189.
Dar, H. A., Zaheer, T., Shehroz, M., Ullah, N., Naz, K., Muhammad, S. A., Zhang, T., & Ali, A. (2019). Immunoinformatics-aided design and evaluation of a potential multi-epitope vaccine against Klebsiella pneumoniae. Vaccines, 7(3), 88.
Moriel, D. G., Beatson, S. A., Wurpel, D. J., Lipman, J., Nimmo, G. R., Paterson, D. L., & Schembri, M. A. (2013). Identification of novel vaccine candidates against multidrug-resistant Acinetobacter baumannii. PLoS ONE, 8(10), e77631.
Solanki, V., & Tiwari, V. (2018). Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against Acinetobacter baumannii. Scientific reports., 8(1), 1–9.
Brunet, Y. R., Hénin, J., Celia, H., & Cascales, E. (2014). Type VI secretion and bacteriophage tail tubes share a common assembly pathway. EMBO reports, 15(3), 315–321.
Jalali, M., Nadoushan, M. J., Rasooli, I., Jahangiri, A., & Dervish, S. (2014). Immunogenicity of amino acid region 7601–8140 in biofilm associated protein of Ab. Pathobiology Research, 16(4), 15–26.
Lin, L., Tan, B., Pantapalangkoor, P., Ho, T., Hujer, A. M., Taracila, M. A., Bonomo, R. A., & Spellberg, B. (2013). Ab rOmpA vaccine dose alters immune polarization and immunodominant epitopes. Vaccine, 31(2), 313–318.
Greco, T. M., & Cristea, I. M. (2017). Proteomics tracing the footsteps of infectious disease. Molecular & Cellular Proteomics, 16(4), S5–S14.
Zheng, M., & Song, L. (2020). Novel antibody epitopes dominate the antigenicity of spike glycoprotein in SARS-CoV-2 compared to SARS-CoV. Cellular & molecular immunology, 17(5), 536–538.
Rakib, A., Sami, S. A., Mimi, N. J., Chowdhury, M. M., Eva, T. A., Nainu, F., Paul, A., Shahriar, A., Tareq, A. M., Emon, N. U., & Chakraborty, S. (2020). Immunoinformatics-guided design of an epitope-based vaccine against severe acute respiratory syndrome coronavirus 2 spike glycoprotein. Computers in biology and medicine, 124, 103967.
Liu, W., & Chen, Y. H. (2005). High epitope density in a single protein molecule significantly enhances antigenicity as well as immunogenicity: A novel strategy for modern vaccine development and a preliminary investigation about B cell discrimination of monomeric proteins. European journal of immunology, 35(2), 505–514.
Blanco, E., Cubillos, C., Moreno, N., Bárcena, J., de la Torre, B.G., Andreu, D., & Sobrino, F. (2013). B epitope multiplicity and B/T epitope orientation influence immunogenicity of foot-and-mouth disease peptide vaccines. Clinical and Developmental Immunology, 2013, 475960.
Clarke, B. E., Newton, S. E., Carroll, A. R., Francis, M. J., Appleyard, G., Syred, A. D., Highfield, P. E., Rowlands, D. J., & Brown, F. (1987). Improved immunogenicity of a peptide epitope after fusion to hepatitis B core protein. Nature, 330(6146), 381–384.
Delany, I., Rappuoli, R., & Seib, K. L. (2013). Vaccines, reverse vaccinology, and bacterial pathogenesis. Cold Spring Harbor perspectives in medicine, 3(5), a012476.
Dalsass, M., Brozzi, A., Medini, D., & Rappuoli, R. (2019). Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Frontiers in immunology, 10, 113.
Moriel, D. G., Beatson, S. A., Wurpel, D. J., Lipman, J., Nimmo, G. R., Paterson, D. L., & Schembri, M. A. (2013). Identification of novel vaccine candidates against multidrug-resistant Ab. PLoS ONE, 8(10), e77631.
Ansari, H., Doosti, A., Kargar, M., Bijanzadeh, M., & Jaafarinia, M. (2018). Cloning of ompA gene from Ab into the eukaryotic expression vector pBudCE4. 1 as DNA vaccine. Indian journal of microbiology, 58(2), 174–181.
Luna, B. M., Yan, J., Reyna, Z., Moon, E., Nielsen, T. B., Reza, H., Lu, P., Bonomo, R., Louie, A., Drusano, G., & Bulitta, J. (2019). Natural history of Ab infection in mice. PLoS ONE, 14(7), e0219824.
Huang, T., Zhao, K., Zhang, Z., Tang, C., Zhang, X., & Yue, B. (2016). DNA vaccination based on pyolysin co-immunized with IL-1β enhances host antibacterial immunity against Trueperella pyogenes infection. Vaccine, 34(30), 3469–3477.
Beiranvand, S., Doosti, A., & Mirzaei, S. A. (2021). Putative novel B-cell vaccine candidates identified by reverse vaccinology and genomics approaches to control Ab serotypes. Infection, Genetics and Evolution., 96, 105138.
Samak T., Gunter D., & Wang Z. (2012). “Prediction of protein solubility in E. coli,” 2012 IEEE 8th International Conference on E-Science, 2012, pp. 1–8, https://doi.org/10.1109/eScience.2012.6404416.
Gui, M., Song, W., Zhou, H., Xu, J., Chen, S., Xiang, Y., & Wang, X. (2017). Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell research., 27(1), 119–129.
Yan, R., Zhang, Y., Li, Y., Xia, L., Guo, Y., & Zhou, Q. (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science, 367(6485), 1444–1448.
Piri-Gharaghie, T. (2021). Polycystic ovary syndrome and genetic factors influencing its development: A review article.
Piri-Gharaghie, T., Jegargoshe-Shirin, N., Saremi-Nouri, S., Khademhosseini, S. H., Hoseinnezhad-Lazarjani, E., Mousavi, A., Kabiri, H., Rajaei, N., Riahi, A., Farhadi-Biregani, A., & Fatehi-Ghahfarokhi, S. (2022). Effects of Imipenem-containing Niosome nanoparticles against high prevalence methicillin-resistant Staphylococcus Epidermidis biofilm formed. Science and Reports, 12(1), 5140. https://doi.org/10.1038/s41598-022-09195-9 PMID: 35332241.
Negahdaripour, M., Nezafat, N., Eslami, M., Ghoshoon, M. B., Shoolian, E., Najafipour, S., Morowvat, M. H., Dehshahri, A., Erfani, N., & Ghasemi, Y. (2018). Structural vaccinology considerations for in silico designing of a multi-epitope vaccine. Infection, Genetics and Evolution, 58, 96–109.
Maleki, A., Russo, G., Parasiliti Palumbo, G. A., & Pappalardo, F. (2021). In silico design of recombinant multi-epitope vaccine against influenza A virus. BMC Bioinformatics, 22(14), 1–19.
Acknowledgements
The authors would like to thank the staff members of the Biotechnology Research Center of Islamic Azad University of Shahrekord Branch in Iran for their help and support.
Author information
Authors and Affiliations
Contributions
Tohid Piri-Gharaghie: Conceptualization, methodology, resources, writing—review and editing, supervision, funding acquisition. Abbas Doosti: Methodology, software, validation, formal analysis, writing—original draft. Seyed Abbas Mirzaei: Visualization, project administration.
Corresponding author
Ethics declarations
Ethical Approval
The study was approved by the Ethics Committee of Islamic Azad University of Shahrekord Branch in Iran [IR.IAU.SHK.REC.1400.046].
Consent to Participate
Not applicable.
Consent to Publish
The authors declare their complete consent to the publication of this manuscript.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Piri-Gharaghie, T., Doosti, A. & Mirzaei, S.A. Identification of Antigenic Properties of Acinetobacter baumannii Proteins as Novel Putative Vaccine Candidates Using Reverse Vaccinology Approach. Appl Biochem Biotechnol 194, 4892–4914 (2022). https://doi.org/10.1007/s12010-022-03995-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-022-03995-5