Abstract
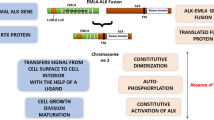
Targeting anaplastic lymphoma kinase (ALK) is one of the important treatment strategies for the treatment of non-small cell lung cancer (NSCLC). In the present perspective, multidimensional approaches were used for the identification of ALK inhibitors. Initially, an e-pharmacophore model was generated using the PHASE algorithm and was used as a 3D query to screen 468,200 molecules of ASINEX database. Prior to the screening process, the model was evaluated for its significance and the ability to differentiate actives from inactives, using enrichment analysis. Subsequently, the hierarchical docking protocol and binding free energy calculations were instigated using GLIDE algorithm and Prime module, respectively. Further, the pharmacokinetic/pharmacodynamics (PK/PD) properties and toxicities of the hit compounds were envisaged respectively using QikProp program, Osiris explorer, and Protox-II algorithm. These approaches retrieved two hits namely BAS 00137817 and BAS 00680055 with acceptable absorption, distribution, metabolism, excretion and toxicity (ADMET) properties and higher affinity towards ALK protein. Additionally, density functional theory calculations and molecular dynamics simulations were performed to validate the inhibitory activity of the lead compounds. It is noteworthy to mention that all the hits constitute of particular scaffolds which play a major role in the downregulation of some ALK-positive lung cancer pathways. We speculate that the outcomes of this research are of substantial prominence in the rational designing of novel and efficacious ALK inhibitors.










Similar content being viewed by others
References
Reck, M., Popat, S., Reinmuth, N., De Ruysscher, D., Kerr, K. M., & Peters, S. (2014). Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology, 25(suppl_3), 27–39.
Stoffel, A. (2010). Targeted therapies for solid tumors. BioDrugs, 24(5), 303–316.
Gerber, D. E. (2008). Targeted therapies: A new generation of cancer treatments. American Family Physician, 77(3), 311–319.
Soda, M., Choi, Y. L., Enomoto, M., Takada, S., Yamashita, Y., Ishikawa, S., Fujiwara, S. I., Watanabe, H., Kurashina, K., Hatanaka, H., & Bando, M. (2007). Identification of the transforming EML4–ALK fusion gene in non-small-cell lung cancer. Nature, 448(7153), 561–566.
Facchinetti, F., Tiseo, M., Di Maio, M., Graziano, P., Bria, E., Rossi, G., & Novello, S. (2016). Tackling ALK in non-small cell lung cancer: the role of novel inhibitors. Translational lung cancer research, 5(3), 301–321.
Soda, M., Takada, S., Takeuchi, K., Choi, Y. L., Enomoto, M., Ueno, T., Haruta, H., Hamada, T., Yamashita, Y., Ishikawa, Y., & Sugiyama, Y. (2008). A mouse model for EML4-ALK-positive lung cancer. Proceedings of the National Academy of Sciences, 105(50), 19893–19897.
Kwak, E. L., Bang, Y. J., Camidge, D. R., Shaw, A. T., Solomon, B., Maki, R. G., Ou, S. H. I., Dezube, B. J., Jänne, P. A., Costa, D. B., & Varella-Garcia, M. (2010). Anaplastic lymphoma kinase inhibition in non–small-cell lung cancer. New England Journal of Medicine, 363(18), 1693–1703.
George, R. E., Sanda, T., Hanna, M., Fröhling, S., Luther II, W., Zhang, J., Ahn, Y., Zhou, W., London, W. B., McGrady, P., & Xue, L. (2008). Activating mutations in ALK provide a therapeutic target in neuroblastoma. Nature, 455(7215), 975–978.
Płużański, A., Piórek, A., & Krzakowski, M. (2012). Crizotinib in the treatment of non-small-cell lung carcinoma. Contemporary Oncology, 16(6), 480–484.
Wu, J., Savooji, J., & Liu, D. (2016). Second-and third-generation ALK inhibitors for non-small cell lung cancer. Journal of Hematology & Oncology, 9(1), 19.
Shaw, A. T., Kim, D. W., Nakagawa, K., Seto, T., Crinó, L., Ahn, M. J., De Pas, T., Besse, B., Solomon, B. J., Blackhall, F., & Wu, Y. L. (2013). Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. New England Journal of Medicine, 368(25), 2385–2394.
Shaw, A. T., Yeap, B. Y., Solomon, B. J., Riely, G. J., Gainor, J., Engelman, J. A., Shapiro, G. I., Costa, D. B., Ou, S. H. I., Butaney, M., & Salgia, R. (2011). Effect of crizotinib on overall survival in patients with advanced non-small-cell lung cancer harbouring ALK gene rearrangement: a retrospective analysis. The Lancet Oncology, 12(11), 1004–1012.
Preethi, B., Shanthi, V., & Ramanathan, K. (2015). Investigation of nalidixic acid resistance mechanism in Salmonella enterica using molecular simulation techniques. Applied Biochemistry and Biotechnology, 177(2), 528–540.
Karthick, V., Shanthi, V., Rajasekaran, R., & Ramanathan, K. (2012). Exploring the cause of oseltamivir resistance against mutant H274Y neuraminidase by molecular simulation approach. Applied Biochemistry and Biotechnology, 167(2), 237–249.
Rohini, K., & Shanthi, V. (2018). Discovery of potent neuraminidase inhibitors using a combination of pharmacophore-based virtual screening and molecular simulation approach. Applied Biochemistry and Biotechnology, 184(4), 1421–1440.
Joung, J. Y., Lee, H. Y., Park, J., Lee, J. Y., Chang, B. H., No, K. T., Nam, K. Y., & Hwang, J. S. (2014). Identification of novel rab27a/melanophilin blockers by pharmacophore-based virtual screening. Applied Biochemistry and Biotechnology, 172(4), 1882–1897.
James, N., & Ramanathan, K. (2018). Ligand-based pharmacophore screening strategy: A pragmatic approach for targeting HER proteins. Applied Biochemistry and Biotechnology, 186(1), 85–108.
Madhulitha, N. R., Pradeep, N., Sandeep, S., Hema, K., & Chiranjeevi, P. (2017). E-pharmacophore model assisted discovery of novel antagonists of nNOS. Biochemistry and Analytical Biochemistry, 6(307), 2161–1009.
Palakurti, R., & Vadrevu, R. (2017). Identification of abelson tyrosine kinase inhibitors as potential therapeutics for Alzheimer’s disease using multiple e-pharmacophore modeling and molecular dynamics. Journal of Biomolecular Structure and Dynamics, 35(4), 883–896.
Clark, D. E., Waszkowycz, B., Wong, M., Lockey, P. M., Adalbert, R., Gilley, J., Clark, J., & Coleman, M. P. (2016). Application of virtual screening to the discovery of novel nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with potential for the treatment of cancer and axonopathies. Bioorganic & Medicinal Chemistry Letters, 26(12), 2920–2926.
Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N., & Bourne, P. E. (2000). The protein data bank. Nucleic Acids Research, 28, 235–242.
Kleywegt, G. J. (2000). Validation of protein crystal structures. Acta Crystallographica Section D: Biological Crystallography, 56(3), 249–265.
Awad, M. M., & Shaw, A. T. (2014). ALK inhibitors in non–small cell lung cancer: Crizotinib and beyond. Clinical advances in hematology & oncology: H&O, 12(7), 429–443.
Sastry, G. M., Adzhigirey, M., Day, T., Annabhimoju, R., & Sherman, W. (2013). Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. Journal of Computer-Aided Molecular Design, 27(3), 221–234.
Shelley, J. C., Cholleti, A., Frye, L. L., Greenwood, J. R., Timlin, M. R., & Uchimaya, M. (2007). Epik: A software program for pK a prediction and protonation state generation for drug-like molecules. Journal of computer-aided molecular design, 21(12), 681–691.
Watts, K. S., Dalal, P., Murphy, R. B., Sherman, W., Friesner, R. A., & Shelley, J. C. (2010). ConfGen: A conformational search method for efficient generation of bioactive conformers. Journal of Chemical Information and Modeling, 50(4), 534–546.
Banks, J. L., Beard, H. S., Cao, Y., Cho, A. E., Damm, W., Farid, R., Felts, A. K., Halgren, T. A., Mainz, D. T., Maple, J. R., & Murphy, R. (2005). Integrated modeling program, applied chemical theory (IMPACT). Journal of Computational Chemistry, 26(16), 1752–1780.
Jorgensen, W. L., & Tirado-Rives, J. (1988). The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. Journal of the American Chemical Society, 110(6), 1657–1666.
Halgren, T. A., Murphy, R. B., Friesner, R. A., Beard, H. S., Frye, L. L., Pollard, W. T., & Banks, J. L. (2004). Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. Journal of Medicinal Chemistry, 47(7), 1750–1759.
Salam, N. K., Nuti, R., & Sherman, W. (2009). Novel method for generating structure-based pharmacophores using energetic analysis. Journal of Chemical Information and Modeling, 49(10), 2356–2368.
Dixon, S. L., Smondyrev, A. M., & Rao, S. N. (2006). PHASE: A novel approach to pharmacophore modeling and 3D database searching. Chemical Biology & Drug Design, 67(5), 370–372.
Truchon, J. F., & Bayly, C. I. (2007). Evaluating virtual screening methods: good and bad metrics for the “early recognition” problem. Journal of chemical information and modeling, 47(2), 488–508.
Sandor, M., Kiss, R., & Keserű, G. M. (2010). Virtual fragment docking by Glide: A validation study on 190 protein−fragment complexes. Journal of Chemical Information and Modeling, 50(6), 1165–1172.
Friesner, R. A., Murphy, R. B., Repasky, M. P., Frye, L. L., Greenwood, J. R., Halgren, T. A., Sanschagrin, P. C., & Mainz, D. T. (2006). Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein−ligand complexes. Journal of Medicinal Chemistry, 49(21), 6177–6196.
Friesner, R. A., Banks, J. L., Murphy, R. B., Halgren, T. A., Klicic, J. J., Mainz, D. T., Repasky, M. P., Knoll, E. H., Shelley, M., Perry, J. K., & Shaw, D. E. (2004). Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. Journal of Medicinal Chemistry, 47(7), 1739–1749.
Lyne, P. D., Lamb, M. L., & Saeh, J. C. (2006). Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MM-GBSA scoring. Journal of Medicinal Chemistry, 49(16), 4805–4808.
Jacobson, M. P., Friesner, R. A., Xiang, Z., & Honig, B. (2002). On the role of the crystal environment in determining protein side-chain conformations. Journal of Molecular Biology, 320(3), 597–608.
Bochevarov, A. D., Harder, E., Hughes, T. F., Greenwood, J. R., Braden, D. A., Philipp, D. M., Rinaldo, D., Halls, M. D., Zhang, J., & Friesner, R. A. (2013). Jaguar: A high-performance quantum chemistry software program with strengths in life and materials sciences. International Journal of Quantum Chemistry, 113(18), 2110–2142.
Pearson, R. G. (1988). Absolute electronegativity and hardness: Application to inorganic chemistry. Inorganic chemistry, 27(4), 734–740.
Parr, R. G., Szentpály, L. V., & Liu, S. (1999). Electrophilicity index. Journal of the American Chemical Society, 121(9), 1922–1924.
Caldwell, G. W. (2000). Compound optimization in early-and late-phase drug discovery: Acceptable pharmacokinetic properties utilizing combined physicochemical, in vitro and in vivo screens. Current Opinion in Drug Discovery & Development, 3(1), 30–41.
Duffy, E. M., & Jorgensen, W. L. (2000). Prediction of properties from simulations: Free energies of solvation in hexadecane, octanol, and water. Journal of the American Chemical Society, 122(12), 2878–2888.
Carrington, C. (2015). Oral targeted therapy for cancer. Australian Prescriber, 38(5), 171–176.
Drwal, M. N., Banerjee, P., Dunkel, M., Wettig, M. R., & Preissner, R. (2014). ProTox: A web server for the in silico prediction of rodent oral toxicity. Nucleic Acids Research, 42(W1), W53–W58.
Hess, B., Kutzner, C., Van Der Spoel, D., & Lindahl, E. (2008). GROMACS 4: Algorithms for highly efficient, load-balanced, and scalable molecular simulation. Journal of Chemical Theory and Computation, 4(3), 435–447.
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A. E., & Berendsen, H. J. (2005). GROMACS: Fast, flexible, and free. Journal of Computational Chemistry, 26(16), 1701–1718.
Scott, W. R., Hünenberger, P. H., Tironi, I. G., Mark, A. E., Billeter, S. R., Fennen, J., Torda, A. E., Huber, T., Krüger, P., & van Gunsteren, W. F. (1999). The GROMOS biomolecular simulation program package. The Journal of Physical Chemistry A, 103(19), 3596–3607.
Daura, X., Mark, A. E., & Van Gunsteren, W. F. (1998). Parametrization of aliphatic CHn united atoms of GROMOS96 force field. Journal of Computational Chemistry, 19(5), 535–547.
Schüttelkopf, A. W., & Van Aalten, D. M. (2004). PRODRG: A tool for high-throughput crystallography of protein–ligand complexes. Acta Crystallographica Section D: Biological Crystallography, 60(8), 1355–1363.
Berendsen, H. J., Postma, J. P., van Gunsteren, W. F., & Hermans, J. (1981). Interaction models for water in relation to protein hydration. In Intermolecular forces (pp. 331–342). Dordrecht: Springer.
Miller III, B. R., & Roitberg, A. E. (2013). Design of e-pharmacophore models using compound fragments for the trans-sialidase of Trypanosoma cruzi: Screening for novel inhibitor scaffolds. Journal of Molecular Graphics and Modelling, 45, 84–97.
Basu, S., & Wallner, B. (2016). Finding correct protein–protein docking models using ProQDock. Bioinformatics, 32(12), i262–i270.
Roskoski Jr., R. (2013). Anaplastic lymphoma kinase (ALK): Structure, oncogenic activation, and pharmacological inhibition. Pharmacological Research, 68(1), 68–94.
Roskoski Jr., R. (2016). Classification of small molecule protein kinase inhibitors based upon the structures of their drug-enzyme complexes. Pharmacological Research, 103, 26–48.
Kakihana, M., Ohira, T., Chan, D., Webster, R. B., Kato, H., Drabkin, H. A., & Gemmill, R. M. (2009). Induction of E-cadherin in lung cancer and interaction with growth suppression by histone deacetylase inhibition. Journal of Thoracic Oncology, 4(12), 1455–1465.
Lee, K. W., Kim, J. H., Park, J. H., Kim, H. P., Song, S. H., Kim, S. G., Kim, T. Y., Jong, H. S., Jung, K. H., Im, S. A., & Kim, T. Y. (2006). Antitumor activity of SK-7041, a novel histone deacetylase inhibitor, in human lung and breast cancer cells. Anticancer Research, 26(5A), 3429–3438.
Mottamal, M., Zheng, S., Huang, T., & Wang, G. (2015). Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules, 20(3), 3898–3941.
Becke, A. D. (1993). Density-functional thermochemistry. III. The role of exact exchange. The Journal of Chemical Physics, 98(5), 5648–5652.
Gill, P. M., Johnson, B. G., Pople, J. A., & Frisch, M. J. (1992). The performance of the Becke—Lee—Yang—Parr (B—LYP) density functional theory with various basis sets. Chemical Physics Letters, 197(4-5), 499–505.
Ramirez-Balderrama, K., Orrantia-Borunda, E., & Flores-Holguin, N. (2017). Calculation of global and local reactivity descriptors of carbodiimides, a DFT study. Journal of Theoretical and Computational Chemistry, 16(03), 1750019.
Pearson, R. G. (1987). Recent advances in the concept of hard and soft acids and bases. Journal of Chemical Education, 64(7), 561.
Shusterman, A. J., & Shusterman, G. P. (1997). Teaching chemistry with electron density models. Journal of Chemical Education, 74(7), 771.
Acknowledgments
The authors are grateful to the Department of Science and Technology-Science and Engineering Research Board (DST-SERB) for funding the research project (File No. EMR/2016/001675) and the management of VIT University, Vellore, for providing the facilities to carry out this work. KR thank ICMR for their support by the International Fellowship for Young Biomedical Scientists Award. VS acknowledges support from Bioinformatics Resources and Applications Facility (BRAF), C-DAC, Pune.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 761 kb)
Rights and permissions
About this article
Cite this article
James, N., Shanthi, V. & Ramanathan, K. Density Functional Theory and Molecular Simulation Studies for Prioritizing Anaplastic Lymphoma Kinase Inhibitors. Appl Biochem Biotechnol 190, 1127–1146 (2020). https://doi.org/10.1007/s12010-019-03156-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03156-1