Abstract
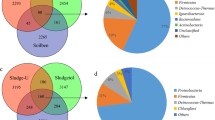
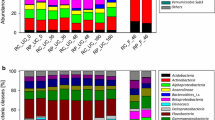
Phenolic compounds are the dominant pollutants in soils contaminated by the coking industry. Ring opening by the hydroxylase gene (bamA) is the key step in the benzoyl-CoA degradation pathway under anaerobic conditions, and a broad spectrum of microorganisms possesses this functional gene, including denitrifiers. The present study analyzed the community structure of denitrifying bacteria and the diversity of the bamA gene for mixed cultures enriched from soil collected at a coking industrial site and then grown under nitrate-reducing conditions on phenol or p-hydroxybenzoate (4HBA), a key intermediate product of anaerobic phenol degradation. Illumina sequencing of the 16S rRNA gene showed different bacterial compositions between the two cultures. The dominant phyla were Proteobacteria, Armatimonadetes, and Planctomycetes in the phenol culture and Proteobacteria and Bacteroidetes in the 4HBA culture. Phylogenetic analysis further demonstrated that bamA genes were associated with four clusters of bacteria, three of known bacteria and one of uncultured bacteria. The diversity of the bamA gene differed from that reported in anaerobic aromatic degradation cultures, suggesting that these enriched cultures may contain new strains unique to coking-contaminated soils. The present study further validates the potential application of this functional gene as a marker for anaerobic biodegradation processes in enrichment cultures from contaminated soil.





Similar content being viewed by others
References
Ahmed, S., Rasul, M. G., Brown, R., & Hashib, M. A. (2011). Influence of parameters on the heterogeneous photocatalytic degradation of pesticides and phenolic contaminants in wastewater: a short review. Journal of Environmental Management, 92(3), 311–330.
Schie, P. M. V., & Young, L. Y. (1998). Isolation and characterization of phenol-degrading denitrifying bacteria. Applied and Environmental Microbiology, 64(7), 2432–2438.
Schink, B., Philipp, B., & Müller, J. (2000). Anaerobic degradation of phenolic compounds. Naturwissenschaften, 87(1), 12–23.
Baek, S. H., Kim, K. H., Yin, C. R., Jeon, C. O., Im, W. T., Kim, K. K., & Lee, S. T. (2003). Isolation and characterization of bacteria capable of degrading phenol and reducing nitrate under low-oxygen conditions. Current Microbiology, 47(6), 462–466.
Li, Z., Suzuki, D., Zhang, C., Yang, S., Nan, J., & Yoshida, N. (2014). Anaerobic 4-chlorophenol mineralization in an enriched culture under iron-reducing conditions. Journal of Bioscience and Bioengineering, 118(5), 529–532.
Narmandakh, A., Gad’On, N., Drepper, F., Knapp, B., Haehnel, W., & Fuchs, G. (2006). Phosphorylation of phenol by phenylphosphate synthase: role of histidine phosphate in catalysis. Journal of Bacteriology, 188(22), 7815–7822.
Schmeling, S., Narmandakh, A., Schmitt, O., Gad’On, N., Schühle, K., & Fuchs, G. (2004). Phenylphosphate synthase: a new phosphotransferase catalyzing the first step in anaerobic phenol metabolism in Thauera aromatica. Journal of Bacteriology, 186(23), 8044–8057.
Schuhle, K., & Fuchs, G. (2004). Phenylphosphate carboxylase: a new C-C lyase involved in anaerobic phenol metabolism in Thauera aromatica. Journal of Bacteriology, 186(14), 4556–4567.
Kuntze, K., Shinoda, Y., Moutakki, H., Mcinerney, M. J., Vogt, C., & Richnow, H. H. (2008). 6-oxocyclohex-1-ene-1-carbonyl-coenzyme A hydrolases from obligately anaerobic bacteria: characterization and identification of its gene as a functional marker for aromatic compounds degrading anaerobes. Environmental Microbiology, 10(6), 1547–1556.
Harwood, C. S., Burchhardt, G., Herrmann, H., & Fuchs, G. (1998). Anaerobic metabolism of aromatic compounds via the benzoyl-coA pathway. FEMS Microbiology Reviews, 22(5), 439–458.
Boll, M., Löffler, C., Morris, B. E. L., & Kung, J. W. (2014). Anaerobic degradation of homocyclic aromatic compounds via arylcarboxyl-coenzyme A esters: organisms, strategies and key enzymes. Environmental Microbiology, 16(3), 612–627.
Porter, A. W., & Young, L. Y. (2013). The bamA gene for anaerobic ring fission is widely distributed in the environment. Frontiers in Microbiology, 4, 302.
Higashioka, Y., Kojima, H., & Fukui, M. (2011). Temperature-dependent differences in community structure of bacteria involved in degradation of petroleum hydrocarbons under sulfate-reducing conditions. Journal of Applied Microbiology, 110(1), 314–322.
Kuntze, K., Vogt, C., Richnow, H. H., & Boll, M. (2011). Combined application of PCR-based functional assays for the detection of aromatic-compound-degrading anaerobes. Applied and Environmental Microbiology, 77(14), 5056–5061.
Staats, M., Braster, M., & Röling, W. F. M. (2011). Molecular diversity and distribution of aromatic hydrocarbon-degrading anaerobes across a landfill leachate plume. Environmental Microbiology, 13(5), 1216–1227.
Li, Y. N., Porter, A. W., Mumford, A., Zhao, X. H., & Young, L. Y. (2012). Bacterial community structure and bamA gene diversity in anaerobic degradation of toluene and benzoate under denitrifying conditions. Journal of Applied Microbiology, 112(2), 269–279.
Porter, A. W., & Young, L. Y. (2014). Benzoyl-CoA, a universal biomarker for anaerobic degradation of aromatic compounds. Advances in Applied Microbiology, 88, 167–203.
Sun, W., Sun, X., & Cupples, A. M. (2014). Presence, diversity and enumeration of functional genes (bssA and bamA) relating to toluene degradation across a range of redox conditions and inoculum sources. Biodegradation, 25(2), 189–203.
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., & Knight, R. (2011). Global patterns of 16s rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences, 108(Suppl 1), 4516–4522.
Evans, P. J., Mang, D. T., Kim, K. S., & Young, L. Y. (1991). Anaerobic degradation of toluene by a denitrifying bacterium. Applied and Environmental Microbiology, 57(4), 1139–1145.
Lane, D. J. (1991). Nucleic acid techniques in bacterial systematic (16S/23S rRNA sequencing) (Vol. 2, pp. 115–175). Chichester: Wiley.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797.
Price, M. N., Dehal, P. S., & Arkin, A. P. (2010). Fasttree 2 – approximately maximum-likelihood trees for large alignments. Plos One, 5, e9490.
Na, J. G., Lee, M. K., Yun, Y. M., Moon, C., Kim, M. S., & Kim, D. H. (2016). Microbial community analysis of anaerobic granules in phenol-degrading UASB by next generation sequencing. Biochemical Engineering Journal, 112, 241–248.
Cheng, C., Zhou, Z., Niu, T., An, Y., Shen, X., Pan, W., Chen, Z. H., & Liu, J. (2017). Effects of side-stream ratio on sludge reduction and microbial structures of anaerobic side-stream reactor coupled membrane bioreactors. Bioresource Technology, 234, 380–388.
van Teeseling, M. C., Mesman, R. J., Kuru, E., Espaillat, A., Cava, F., Brun, Y. V., VanNieuwenhze, M. S., Kartal, B., & van Niftrik, L. (2015). Anammox planctomycetes have a peptidoglycan cell wall. Nature Communications, 6(6878), 6878.
Ma, Q., Qu, Y., Shen, W., Zhang, Z., Wang, J., Liu, Z., Li, D., Li, H., & Zhou, J. (2014). Bacterial community compositions of coking wastewater treatment plants in steel industry revealed by Illumina high-throughput sequencing. Bioresource Technology, 179, 436–443.
Ren, L. F., Chen, R., Zhang, X. F., Shao, J. H., & He, Y. L. (2017). Phylotypes belonging to the Bacteroidetes are known to be physiologically diverse and are found in a wide range of environments, e.g. soil, freshwater, ocean and the human gastrointestinal tract. Bioresource Technology, 244, 1121–1128.
Ibarbalz, F. M., Figuerola, E. L., & Erijman, L. (2013). Industrial activated sludge exhibit unique bacterial community composition at high taxonomic ranks. Water Research, 47(11), 3854–3864.
Manefield, M., Griffiths, R. I., Leigh, M. B., Fisher, R., & Whiteley, A. S. (2005). Functional and compositional comparison of two activated sludge communities remediating coking effluent. Environmental Microbiology, 7(5), 715–722.
Mechichi, T., Stackebrandt, E., Gad’On, N., & Fuchs, G. (2002). Phylogenetic and metabolic diversity of bacteria degrading aromatic compounds under denitrifying conditions, and description of Thauera phenylacetica sp. nov., Thauera aminoaromatica sp. nov., and Azoarcus buckelii sp. nov. Archives of Microbiology, 178(1), 26–35.
Morgan-Sagastume, F., Nielsen, J. L., & Nielsen, P. H. (2008). Substrate-dependent denitrification of abundant probe-defined denitrifying bacteria in activated sludge. FEMS Microbiology Ecology, 66(2), 447–461.
Lee, D. J., Wong, B. T., & Adav, S. S. (2014). Azoarcus taiwanensis, sp. nov. a denitrifying species isolated from a hot spring. Applied Microbiology and Biotechnology, 98(3), 1301–1307.
Vedler, E., Heinaru, E., Jutkina, J., Viggor, S., Koressaar, T., Remm, M., & Heinaru, A. (2013). Limnobacter spp. as newly detected phenol-degraders among Baltic Sea surface water bacteria characterised by comparative analysis of catabolic genes. Systematic and Applied Microbiology, 36(8), 525–532.
Sass, A. M., Schmerk, C., Agnoli, K., Norville, P. J., Eberl, L., Valvano, M. A., & Mathenthiralingam, E. (2013). The unexpected discovery of a novel low-oxygen-activated locus for the anoxic persistence of Burkholderia cenocepacia. The ISME Journal, 7(8), 1568–1581.
Mi, W., Zhao, J., Ding, X., Ge, G., & Zhao, R. (2017). Treatment performance, nitrous oxide production and microbial community under low-ammonium wastewater in a CANON process. Water Science and Technology, 76(12), 3468–3477.
Wang, Z., Yang, Y. Y., Dai, Y., & Xie, S. G. (2015). Anaerobic biodegradation of nonylphenol in river sediment under nitrate- or sulfate-reducing conditions and associated bacterial community. Journal of Hazardous Materials, 286, 306–314.
Bai, N. (2018) Microbiological study on the formation of black and smelly water in typical areas. Master thesis, Capital University of Economics and Business, Beijing, China.
Andreoni, V., Cavalca, L., Rao, M. A., Nocerino, G., Bernasconi, S., Dell’Amico, E., Colombo, M., & Gianfreda, L. (2004). Bacterial communities and enzyme activities of PAHs polluted soils. Chemosphere, 57(5), 401–412.
Huang, Y., Hou, X., Liu, S., & Ni, J. (2016). Correspondence analysis of bio-refractory compounds degradation and microbiological community distribution in anaerobic filter for coking wastewater treatment. Chemical Engineering Journal, 304, 864–872.
Sampaio, D. S., Almeida, J. R. B., Jesus, H. E. D., Rosado, A. S., & Jurelevicius, D. (2017). Distribution of anaerobic hydrocarbon-degrading bacteria in soils from King George island, Maritime Antarctica. Microbial Ecology, 74(24), 1–11.
Ruan, M. Y., Liang, B., Mbadinga, S. M., Zhou, L., Wang, L. Y., Liu, J. F., Gu, J. D., & Mu, B. Z. (2016). Molecular diversity of bacterial bamA gene involved in anaerobic degradation of aromatic hydrocarbons in mesophilic petroleum reservoirs. International Biodeterioration & Biodegradation, 114, 122–128.
Lahme, S., Harder, J., & Rabus, R. (2012). Anaerobic degradation of 4-methylbenzoate by a newly isolated denitrifying bacterium, strain pmbn1. Applied and Environmental Microbiology, 78(5), 1606–1610.
Wirth, B., Krebs, M., & Andert, J. (2015). Anaerobic degradation of increased phenol concentrations in batch assays. Environmental Science and Pollution Research, 22(23), 19048–19059.
Acknowledgments
Jianwei Wang is acknowledged for the helpful discussion in the design of the study and for the help with culture setting up. Jin Liu and Jingyi Fu are both thanked for collecting soil samples from the coking plant.
Funding
This study was supported by the Science and Technology Planning Project of Guangdong Province (No. 2017B030314092), Open Fund of Key Laboratory of Eco-geochemistry, Ministry of Natural Resources (No. ZSDHJJ201804), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (No. 2016146, 201802026), the Scientific and Technological Project of Shanxi Province (No. 2015021119), and the National Natural Science Foundation of China (No. 51378330, 51408396).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 13 kb)
Rights and permissions
About this article
Cite this article
Li, Y., Li, J., Wang, D. et al. Denitrifying Microbial Community Structure and bamA Gene Diversity of Phenol Degraders in Soil Contaminated from the Coking Process. Appl Biochem Biotechnol 190, 966–981 (2020). https://doi.org/10.1007/s12010-019-03144-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-019-03144-5