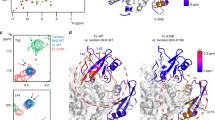
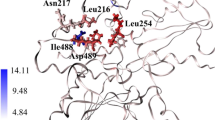
Abstract
There is currently considerable interest in SHP2 as a potential target for treatment of cancer. Mutation in SHP2, particularly the E76A mutation, has been found to seriously confer the phosphatase high activity. Recently, two compounds, 1 and 23, have been reported as potent allosteric inhibitors of both SHP2 wild type (SHP2WT) and the E76A mutant (SHP2E76A), with higher activity than other inhibitors. However, the structural and dynamic implications of their inhibitory mechanisms are yet unexplored which deserve further attention. Herein, the MD simulation applies to gain insight into the atomistic nature of each binding mode of inhibitors 1 and 23 in both SHP2WT and SHP2E76A. The comparative analysis reveals inhibitor 1 can freeze SHP2WT and SHP2E76A in their auto-inhibited conformation better than 23, in agreement with experimental data. GLU250 in both SHP2WT and SHP2E76A and ARG111 and ARG229 in SHP2E76A play a crucial role in the higher activity of 1 compared to 23. The mutation E76A increases the binding affinity of 1 and 23 compared to the wild type, implying that the two inhibitors have been well adopted by the E76A mutant. The findings here can substantially shed light on new strategies for developing novel classes of SHP2 inhibitors with increased potency.













Similar content being viewed by others
References
Tonks, N. K. (2006). Protein tyrosine phosphatases: from genes, to function, to disease. Nature Reviews Molecular Cell Biology, 7(11), 833–846.
Van Huijsduijnen, R. H., Bombrun, A., & Swinnen, D. (2002). Selecting protein tyrosine phosphatases as drug targets. Drug Discovery Today, 7(19), 1013–1019.
Kollman, P. A., Massova, I., Reyes, C., Kuhn, B., Huo, S., Chong, L., Lee, M., Lee, T., Duan, Y., & Wang, W. (2000). Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Accounts of Chemical Research, 33(12), 889–897.
Zhang, Z.-Y. (2001). Protein tyrosine phosphatases: prospects for therapeutics. Current Opinion in Chemical Biology, 5(4), 416–423.
Bialy, L., & Waldmann, H. (2005). Inhibitors of protein tyrosine phosphatases: next-generation drugs? Angewandte Chemie International Edition, 44(25), 3814–3839.
Mohi, M. G., & Neel, B. G. (2007). The role of Shp2 (PTPN11) in cancer. Current Opinion in Genetics & Development, 17(1), 23–30.
Chan, R. J., & Feng, G.-S. (2007). PTPN11 is the first identified proto-oncogene that encodes a tyrosine phosphatase. Blood, 109(3), 862–867.
Matozaki, T., Murata, Y., Saito, Y., Okazawa, H., & Ohnishi, H. (2009). Protein tyrosine phosphatase SHP-2: a proto-oncogene product that promotes Ras activation. Cancer Science, 100(10), 1786–1793.
Gavrieli, M., Watanabe, N., Loftin, S. K., Murphy, T. L., & Murphy, K. M. (2003). Characterization of phosphotyrosine binding motifs in the cytoplasmic domain of B and T lymphocyte attenuator required for association with protein tyrosine phosphatases SHP-1 and SHP-2. Biochemical and Biophysical Research Communications, 312(4), 1236–1243.
Yokosuka, T., Takamatsu, M., Kobayashi-Imanishi, W., Hashimoto-Tane, A., Azuma, M., & Saito, T. (2012). Programmed cell death 1 forms negative costimulatory microclusters that directly inhibit T cell receptor signaling by recruiting phosphatase SHP2. Journal of Experimental Medicine, 209(6), 1201–1217.
Chemnitz, J. M., Parry, R. V., Nichols, K. E., June, C. H., & Riley, J. L. (2004). SHP-1 and SHP-2 associate with immunoreceptor tyrosine-based switch motif of programmed death 1 upon primary human T cell stimulation, but only receptor ligation prevents T cell activation. The Journal of Immunology, 173(2), 945–954.
Darian, E., Guvench, O., Yu, B., Qu, C. K., & MacKerell, A. D. (2011). Structural mechanism associated with domain opening in gain-of-function mutations in SHP2 phosphatase. Proteins: Structure, Function, and Bioinformatics, 79(5), 1573–1588.
Hof, P., Pluskey, S., Dhe-Paganon, S., Eck, M. J., & Shoelson, S. E. (1998). Crystal structure of the tyrosine phosphatase SHP-2. Cell, 92(4), 441–450.
Xie, J., Si, X., Gu, S., Wang, M., Shen, J., Li, H., Shen, J., Li, D., Fang, Y., & Liu, C. (2017). Allosteric inhibitors of SHP2 with therapeutic potential for cancer treatment. Journal of Medicinal Chemistry, 60(24), 10205–10219.
Garcia Fortanet, J., Chen, C. H.-T., Chen, Y.-N. P., Chen, Z., Deng, Z., Firestone, B., Fekkes, P., Fodor, M., Fortin, P. D., & Fridrich, C. (2016). Allosteric inhibition of SHP2: identification of a potent, selective, and orally efficacious phosphatase inhibitor. Journal of Medicinal Chemistry, 59(17), 7773–7782.
Butterworth, S., Overduin, M., & Barr, A. J. (2014). Targeting protein tyrosine phosphatase SHP2 for therapeutic intervention. Future Medicinal Chemistry, 6(12), 1423–1437.
M Scott, L., R Lawrence, H., M Sebti, S., J Lawrence, N., & Wu, J. (2010). Targeting protein tyrosine phosphatases for anticancer drug discovery. Current Pharmaceutical Design, 16(16), 1843–1862.
Song, K., Liu, X., Huang, W., Lu, S., Shen, Q., Zhang, L., & Zhang, J. (2017). Improved method for the identification and validation of allosteric sites. Journal of Chemical Information and Modeling, 57(9), 2358–2363.
Boycott, K. M., Rath, A., Chong, J. X., Hartley, T., Alkuraya, F. S., Baynam, G., Brookes, A. J., Brudno, M., Carracedo, A., & den Dunnen, J. T. (2017). International cooperation to enable the diagnosis of all rare genetic diseases. The American Journal of Human Genetics, 100(5), 695–705.
Huang, W., Wang, G., Shen, Q., Liu, X., Lu, S., Geng, L., Huang, Z., & Zhang, J. (2015). ASBench: benchmarking sets for allosteric discovery. Bioinformatics, 31(15), 2598–2600.
De Vivo, M., Masetti, M., Bottegoni, G., & Cavalli, A. (2016). Role of molecular dynamics and related methods in drug discovery. Journal of Medicinal Chemistry, 59(9), 4035–4061.
Ma, X., Meng, H., & Lai, L. (2016). Motions of allosteric and orthosteric ligand-binding sites in proteins are highly correlated. Journal of Chemical Information and Modeling, 56(9), 1725–1733.
Salamoun, J. M., & Wipf, P. (2016). Allosteric modulation of phosphatase activity may redefine therapeutic value. Washington, DC: ACS Publications.
Chio, C. M., Lim, C. S., & Bishop, A. C. (2015). Targeting a cryptic allosteric site for selective inhibition of the oncogenic protein tyrosine phosphatase Shp2. Biochemistry, 54(2), 497–504.
Ahmed-Belkacem, A., Guichou, J.-F., Brillet, R., Ahnou, N., Hernandez, E., Pallier, C., & Pawlotsky, J.-M. (2014). Inhibition of RNA binding to hepatitis C virus RNA-dependent RNA polymerase: a new mechanism for antiviral intervention. Nucleic Acids Research, 42(14), 9399–9409.
Miyamoto, D., Miyamoto, M., Takahashi, A., Yomogita, Y., Higashi, H., Kondo, S., & Hatakeyama, M. (2008). Isolation of a distinct class of gain-of-function SHP-2 mutants with oncogenic RAS-like transforming activity from solid tumors. Oncogene, 27(25), 3508–3515.
Chan, G., Kalaitzidis, D., & Neel, B. G. (2008). The tyrosine phosphatase Shp2 (PTPN11) in cancer. Cancer and Metastasis Reviews, 27(2), 179–192.
Tartaglia, M., Mehler, E., Goldberg, R., Zampino, G., Brunner, H., Kremer, H., van der Burgt, I., Crosby, A., Ion, A., & Jeffery, S. (2001). Mutations in the protein tyrosine kinase gene, PTPN11, cause Noonan syndrome. Nature Genetics, 29(4), 491.
Bentires-Alj, M., Paez, J. G., David, F. S., Keilhack, H., Halmos, B., Naoki, K., Maris, J. M., Richardson, A., Bardelli, A., & Sugarbaker, D. J. (2004). Activating mutations of the Noonan syndrome-associated SHP2/PTPN11 gene in human solid tumors and adult acute myelogenous leukemia. Cancer Research, 64(24), 8816–8820.
Che, X., Du, X.-X., Cai, X., Zhang, J., Xie, W. J., Long, Z., Ye, Z.-Y., Zhang, H., Yang, L., & Su, X.-D. (2017). Single mutations reshape the structural correlation network of the DMXAA–human STING complex. The Journal of Physical Chemistry B, 121(9), 2073–2082.
Aier, I., Varadwaj, P. K., & Raj, U. (2016). Structural insights into conformational stability of both wild-type and mutant EZH2 receptor. Scientific Reports, 6(1), 34984.
Bai, Q., Pérez-Sánchez, H., Zhang, Y., Shao, Y., Shi, D., Liu, H., & Yao, X. (2014). Ligand induced change of β 2 adrenergic receptor from active to inactive conformation and its implication for the closed/open state of the water channel: insight from molecular dynamics simulation, free energy calculation and Markov state model analysis. Physical Chemistry Chemical Physics, 16(30), 15874–15885.
Bhakat, S., Martin, A. J., & Soliman, M. E. (2014). An integrated molecular dynamics, principal component analysis and residue interaction network approach reveals the impact of M184V mutation on HIV reverse transcriptase resistance to lamivudine. Molecular BioSystems, 10(8), 2215–2228.
Eswar, N., Webb, B., Marti-Renom, M. A., Madhusudhan, M., Eramian, D., Shen, M. y., Pieper, U., & Sali, A. (2006). Comparative protein structure modeling using Modeller. Current Protocols in Bioinformatics, 15(1), 5.6. 1–5.6. 30.
Becke, A. D. (1993). Density-functional thermochemistry. III. The role of exact exchange. The Journal of Chemical Physics, 98(7), 5648–5652.
Frisch, M., Trucks, G., Schlegel, H., Scuseria, G., Robb, M., Cheeseman, J., Scalmani, G., Barone, V., Petersson, G., Nakatsuji, H. (2016). Gaussian Inc. 16, revision A. 03; Gaussian Inc. Wallingford, CT.
Pettersen, E. F., Goddard, T. D., Huang, C. C., Couch, G. S., Greenblatt, D. M., Meng, E. C., & Ferrin, T. E. (2004). UCSF chimera—a visualization system for exploratory research and analysis. Journal of Computational Chemistry, 25(13), 1605–1612.
Ramharack, P., Oguntade, S., & Soliman, M. E. (2017). Delving into Zika virus structural dynamics—a closer look at NS3 helicase loop flexibility and its role in drug discovery. RSC Advances, 7(36), 22133–22144.
Olotu, F. A., Agoni, C., Adeniji, E., Abdullahi, M., & Soliman, M. E. (2018). Probing Gallate-mediated selectivity and high-affinity binding of epigallocatechin Gallate: a way-forward in the design of selective inhibitors for anti-apoptotic Bcl-2 proteins. Applied Biochemistry and Biotechnology, 1–20.
Ncub, B. N., Ramharack, P., & Soliman, M. E. (2017). An “all-in-one” pharmacophoric architecture for the discovery of potential broad-spectrum anti-flavivirus drugs. Applied Biochemistry and Biotechnology, 1–16.
Ndagi, U., Mhlongo, N. N., & Soliman, M. E. (2017). Emergence of a promising lead compound in the treatment of triple negative breast cancer: an insight into conformational features and ligand binding landscape of c-Src protein with UM-164. Applied Biochemistry and Biotechnology, 185, 655–675.
Srivastava, H. K., & Sastry, G. N. (2012). Molecular dynamics investigation on a series of HIV protease inhibitors: assessing the performance of MM-PBSA and MM-GBSA approaches. Journal of Chemical Information and Modeling, 52(11), 3088–3098.
Mathew, B., Adeniyi, A. A., Dev, S., Joy, M., Ucar, G. l., Mathew, G. E., Singh-Pillay, A., & Soliman, M. E. (2017). Pharmacophore-based 3D-QSAR analysis of thienyl chalcones as a new class of human MAO-B inhibitors: investigation of combined quantum chemical and molecular dynamics approach. The Journal of Physical Chemistry B, 121(6), 1186–1203.
Case, D. A., Cheatham, T. E., Darden, T., Gohlke, H., Luo, R., Merz, K. M., Onufriev, A., Simmerling, C., Wang, B., & Woods, R. J. (2005). The amber biomolecular simulation programs. Journal of Computational Chemistry, 26(16), 1668–1688.
Berendsen, H. J., Postma, J. v., van Gunsteren, W. F., DiNola, A., & Haak, J. (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics, 81(8), 3684–3690.
Roe, D. R., & Cheatham III, T. E. (2013). PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. Journal of Chemical Theory and Computation, 9(7), 3084–3095.
Seifert, E. (2014). OriginPro 9.1: scientific data analysis and graphing software—software review. ACS Publications
Tsui, V., & Case, D. A. (2000). Theory and applications of the generalized Born solvation model in macromolecular simulations. Biopolymers, 56(4), 275–291.
Massova, I., & Kollman, P. A. (2000). Combined molecular mechanical and continuum solvent approach (MM-PBSA/GBSA) to predict ligand binding. Perspectives in Drug Discovery and Design, 18(1), 113–135.
Barreca, M. L., Lee, K. W., Chimirri, A., & Briggs, J. M. (2003). Molecular dynamics studies of the wild-type and double mutant HIV-1 integrase complexed with the 5CITEP inhibitor: mechanism for inhibition and drug resistance. Biophysical Journal, 84(3), 1450–1463.
Harvey, M., & De Fabritiis, G. (2009). An implementation of the smooth particle mesh Ewald method on GPU hardware. Journal of Chemical Theory and Computation, 5(9), 2371–2377.
Word, J. M., Lovell, S. C., LaBean, T. H., Taylor, H. C., Zalis, M. E., Presley, B. K., Richardson, J. S., & Richardson, D. C. (1999). Visualizing and quantifying molecular goodness-of-fit: small-probe contact dots with explicit hydrogen atoms1. Journal of Molecular Biology, 285(4), 1711–1733.
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., Amin, N., Schwikowski, B., & Ideker, T. (2003). Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504.
Doncheva, N. T., Klein, K., Domingues, F. S., & Albrecht, M. (2011). Analyzing and visualizing residue networks of protein structures. Trends in Biochemical Sciences, 36(4), 179–182.
Yu, Z.-H., Zhang, R.-Y., Walls, C. D., Chen, L., Zhang, S., Wu, L., Liu, S., & Zhang, Z.-Y. (2014). Molecular basis of gain-of-function LEOPARD syndrome-associated SHP2 mutations. Biochemistry, 53(25), 4136–4151.
Acknowledgements
The authors acknowledge the School of Health Sciences, the University of KwaZulu-Natal, Westville Campus, for financial assistance. They also acknowledge the Centre for High Performance Computing (CHPC, www.chpc.ac.za), Cape Town, South Africa, for computational resources. The editorial support of Ms. Carrin Martin, editor for the School of Health Sciences at UKZN, is also acknowledged.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 25 kb)
Rights and permissions
About this article
Cite this article
Farrokhzadeh, A., Akher, F.B. & Soliman, M.E.S. Probing the Dynamic Mechanism of Uncommon Allosteric Inhibitors Optimized to Enhance Drug Selectivity of SHP2 with Therapeutic Potential for Cancer Treatment. Appl Biochem Biotechnol 188, 260–281 (2019). https://doi.org/10.1007/s12010-018-2914-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-018-2914-0