Abstract
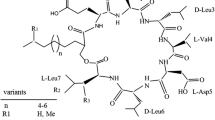
The lipopeptide and its homologues are a kind of the five major biosurfactants with prominent interfacial and biological activities. A suite of mutagenesis method was adopted to expose a wild lipopeptide-producing strain Bacillus subtilis HSO121 to improve lipopeptide yield, and a stable mutant named R2-104 with a 2.0-fold production of lipopeptide was obtained. Compared to that of the wild strain HSO121, the lipopeptide produced by R2-104 showed a similar surface activity, but the course profiles of lipopeptide production during cultivation were different, with the peak yield of 500 mg at about 9 h by R2-104, and 400 mg at about 5 h by HSO121. The constituent abundance of the lipopeptide homologues produced by R2-104 was also different from that by HSO121. Combined methods of ESI-MS, GC-MS and MS-MS were applied for structural characterization of lipopeptide homologues, and it showed that the lipopeptides produced by R2-104 and HSO121 were attributed to a surfactin family with different constituents. The dominant constituent of the surfactin family produced by R2-104 was anteiso C15-surfactin with a relative content of 43.8 %, while the dominant one produced by HSO121was iso C14-surfactin with a relative content of 33.1 %.






Similar content being viewed by others
References
Kim, P. I., Bai, H., Bai, D., Chae, H., Chung, S., Kim, Y., et al. (2004). Purification and characterization of a lipopeptide produced by Bacillus thuringiensis CMB26. Journal of Applied Microbiology, 97, 942–949.
Bezza, F. A., & Chirwa, E. M. N. (2015). Production and applications of lipopeptide biosurfactant for bioremediation and oil recovery by Bacillus subtilis CN2. Biochemical Engineering Journal, 101, 168–178.
Raaijmakers, J. M., Bruijn, I. D., & Kock, M. J. D. D. (2006). Cyclic lipopeptide production by plant-associated Pseudomonas spp.: diversity, activity, biosynthesis, and regulation. Molecular Plant-Microbe Interactions, 19(7), 699–710.
Trelita, D. S., & Saroj, B. (2012). Isolation and characterization of a lipopeptide bioemulsifier produced by Pseudomonas nitroreducens TSB.MJ10 isolated from a mangrove ecosystem. Bioresource Technology, 123, 256–262.
Souza, J. T. D., Boer, M. D., Waard, P. D., Beek, T. A. V., & Raaijmakers, J. M. (2003). Biochemical, genetic, and zoosporicidal properties of cyclic lipopeptide surfactants produced by Pseudomonas fluorescens. Applied and Environmental Microbiology, 69(12), 7161–7172.
Banat, I. M., Franzetti, A., Gandolfi, I., Bestetti, G., Martinotti, M. G., Fracchia, L., et al. (2010). Microbial biosurfactants production, applications and future potential. Applied Microbiology and Biotechnology, 87(2), 427–444.
Morikawa, M., Hirata, Y., & Imanaka, T. (2000). A study on the structure function relationship of lipopeptide biosurfactants. Biochimica et Biophysica Acta, 1488, 211–218.
Sachdev, D. P., & Cameotra, S. S. (2013). Biosurfactants in agriculture. Applied Microbiology and Biotechnology, 97(3), 1005–1016.
Chen, L. L., Wang, N., Wang, X. M., Hu, J. C., & Wang, S. J. (2010). Characterization of two anti-fungal lipopeptides produced by Bacillus amyloliquefaciens SH-B10. Bioresource Technology, 101(22), 8822–8827.
Weis, F., Beiras-Fernandez, A., & Schelling, G. (2008). Daptomycin, a lipopeptide antibiotic in clinical practice. Current Opinion in Investigational Drugs, 9(8), 879–884.
BenMohamed, L., Wechsler, S. L., Nesburn, A. B., BenMohamed, L., Wechsler, S. L., & Nesburn, A. B. (2002). Lipopeptide vaccines—yesterday, today, and tomorrow. The Lancet Infectious Diseases, 2(7), 425–431.
Nitschke, M., & Costa, S. G. V. A. O. (2007). Biosurfactants in food industry. Trends in Food Science & Technology, 18(5), 252–259.
Mulligan, C. N. (2009). Recent advances in the environmental applications of biosurfactants. Current Opinion in Colloid & Interface Science, 14(5), 372–378.
Pereira, J. F. B., Gudiña, E. J., Costa, R., Vitorino, R., Teixeira, J. A., Coutinho, J. A. P., et al. (2013). Optimization and characterization of biosurfactant production by Bacillus subtilis isolates towards microbial enhanced oil recovery applications. Fuel, 111, 259–268.
Peypoux, F., Bonmatin, J. M., & Wallach, J. (1999). Recent trends in the biochemistry of surfactin. Applied Microbiology and Biotechnology, 51(5), 553–563.
Mizumoto, S., Hirai, M., & Shoda, M. (2006). Production of lipopeptide antibiotic iturin A using soybean curd residue cultivated with Bacillus subtilis in solid-state fermentation. Applied Microbiology and Biotechnology, 72(5), 869–875.
Honma, M., Tanaka, K., Konno, K., Tsuge, K., Okuno, T., & Hashimoto, M. (2012). Termination of the structural confusion between plipastatin A1 and fengycin IX. Bioorganic & Medicinal Chemistry, 20(12), 3793–3798.
Abderrahmani, A., Tapi, A., Nateche, F., Chollet, M., Leclère, V., Wathelet, B., et al. (2011). Bioinformatics and molecular approaches to detect NRPS genes involved in the biosynthesis of kurstakin from Bacillus thuringiensis. Applied Microbiology and Biotechnology, 92(3), 571–581.
Hajfarajollah, H., Mokhtarani, B., & Noghabi, K. A. (2014). Newly antibacterial and antiadhesive lipopeptide biosurfactant secreted by a probiotic strain, Propionibacterium freudenreichii. Applied Biochemistry and Biotechnology, 174(8), 2725–2740.
Shaligram, N. S., & Singhal, R. S. (2010). Surfactin—a review on biosynthesis, fermentation, purification and applications. Food Technology and Biotechnology, 42(2), 119–134.
Das, K., & Mukherjee, A. K. (2007). Comparison of lipopeptide biosurfactants production by Bacillus subtilis strains in submerged and solid state fermentation systems using a cheap carbon source: Some industrial applications of biosurfactants. Process Biochemistry, 42(8), 1191–1199.
Piedrahita-Aguirre, C. A., Bastos, R. G., Carvalho, A. L., & Alegre, R. M. (2014). The influence of process parameters in production of lipopeptide iturin A using aerated packed bed bioreactors in solid-state fermentation. Bioprocess and Biosystems Engineering, 37(8), 1569–1576.
Miao, V., Coeffet-Le, G. M. F., Nguyen, K., Brian, P., Penn, J., Whiting, A., et al. (2006). Genetic engineering in Streptomyces roseosporus to produce hybrid lipopeptide antibiotics. Chemistry & Biology, 13(3), 269–276.
Li, Y. X., Li, Z. R., Yamanaka, K., Xu, Y., Zhang, W. P., Vlamakis, H., et al. (2015). Directed natural product biosynthesis gene cluster capture and expression in the model bacterium Bacillus subtilis. Scientific Reports, 5, 9383.
Qiu, Y. M., Xiao, F., Wei, X. T., Wen, Z. Y., & Chen, S. W. (2014). Improvement of lichenysin production in Bacillus licheniformis by replacement of native promoter of lichenysin biosynthesis operon and medium optimization. Applied Microbiology and Biotechnology, 98(21), 8895–8903.
Connor, M. R., Cann, A. F., & Liao, J. C. (2010). 3-Methyl-1-butanol production in Escherichia coli: random mutagenesis and two-phase fermentation. Applied Microbiology and Biotechnology, 86(4), 1155–1164.
Lotfy, W. A., Ghanem, K. M., & El-Helow, E. R. (2007). Citric acid production by a novel Aspergillus niger isolate: I. Mutagenesis and cost reduction studies. Bioresource Technology, 98(18), 3464–3469.
Himabindu, M., Potumarthi, R., & Jetty, A. (2007). Enhancement of gentamicin production by mutagenesis and non-nutritional stress conditions in Micromonospora echinospora. Process Biochemistry, 42(9), 1352–1356.
Kanda, M., Tsuboi, M., Sakamoto, K., Shimizu, S., Yamashita, M., & Honda, H. (2009). Improvement of FR901379 production by mutant selection and medium optimization. Journal of Bioscience and Bioengineering, 107(5), 530–534.
Chen, H., Chen, Z. J., Wu, M. B., & Deng, S. X. (2010). Screening the Fusarium graminearum inhibitory mutant strain from Bacillus subtilis by atmospheric-pressure plasma jet. Journal of Applied Microbiology, 108(1), 96–103.
Wu, C. J., Li, C. W., & Cui, C. B. (2014). Seven new and two known lipopeptides as well as five known polyketides: the activated production of silent metabolites in a marine-derived fungus by chemical mutagenesis strategy using diethyl sulphate. Marine Drugs, 12(4), 1815–1838.
Yang, S. Z., Wei, D. Z., & Mu, B. Z. (2007). Determination of the structure of the fatty acid chain in a cyclic lipopeptide using GC-MS. Journal of Biochemical and Biophysical Methods, 70(3), 519–523.
Yakimov, M. M., Abraham, W.-R., Meyer, H., Giuliano, L., & Golyshin, P. N. (1999). Structural characterization of lichenysin A components by fast atom bombardment tandem mass spectrometry. Biochimica et Biophysica Acta, 1438(2), 273–280.
Yang, S. Z., Wei, D. Z., & Mu, B. Z. (2006). Determination of the amino acid sequence in a cyclic lipopeptide using MS with DHT mechanism. Journal of Biochemical and Biophysical Methods, 68(1), 69–74.
Lv, Y. N., Yang, S. Z., & Mu, B. Z. (2005). Isolation and identification of a lipopeptide. Microbiology, 32(1), 67–73.
Mukherjee, S., Das, P., & Sen, R. (2009). Rapid quantification of a microbial surfactant by a simple turbidometric method. Journal of Microbiological Methods, 76, 38–42.
You, J., Yang, S. Z., & Mu, B. Z. (2015). Structural characterization of lipopeptides from Enterobacter sp. strain N18 reveals production of surfactin homologues. European Journal of Lipid Science and Technology, 117(6), 890–898.
Zhao, Y., Yang, S. Z., & Mu, B. Z. (2012). Quantitative analyses of the isoforms of surfactin produced by Bacillus subtilis HSO 121 using GC-MS. Analytical Sciences, 28, 789–793.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 21203063 and 51574125) and the 863 Program (Grant No. 2013AA064403).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 669 kb)
Rights and permissions
About this article
Cite this article
Meng, Y., Zhao, W., You, J. et al. Structural Analysis of the Lipopeptide Produced by the Bacillus subtilis Mutant R2-104 with Mutagenesis. Appl Biochem Biotechnol 179, 973–985 (2016). https://doi.org/10.1007/s12010-016-2044-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-016-2044-5