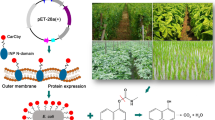
Abstract
Compounds including organophosphorus pesticides (OPs) and chemical nerve agents are toxic compounds synthesized recently which disrupt the mechanisms of neural transmission. Therefore, a critical requirement is the development of a bio-refining technology to facilitate the biodegradation of organophosphorus pollutants. The diisopropylfluorophosphatase (DFPase, EC 3.1.8.2) from the ganglion and brain of Loligo vulgaris acts on P–F bonds present in some OPs. Intracellular production of OPs-degrading enzymes or the use of native bacteria and fungi leads to a low degradation rate of OPs due to a mass transfer issue which reduces the overall catalytic efficiency. To overcome this challenge, we expressed DFPase on the surface of E. coli for the first time by employing the N-terminal domain of the ice nucleation protein (InaV-N) as an anchoring motif. Tracking the recombinant protein confirmed that DFPase is successfully located on the outer membrane. Further studies on its activity to degrade diisopropylfluorophosphate (DFP) showed its significant ability for the biodegradation of diisopropylfluorophosphate (DFP) with a specific activity of 500 U/mg of wet cell weight. Recombinant cells could also degrade chlorpyrifos (Cp) with an activity equivalent to a maximum value of 381.44 U/ml with a specific activity of 476.75 U/mg of cell, analyzed using HPLC technique. The optimum activity of purified DFPase was found at 30 °C. A more increased activity was also obtained in the presence of glucose-mineral-salt (GMS) supplemented with tryptone and 100 mg/L Co2+ ion. These results highlight the high potential of the InaV-N anchoring domain to produce an engineered bacterium that can be used in the bioremediation of pesticide-contaminated environments.








Similar content being viewed by others
References
Balali-Mood, M., & Saber, H. (2012). Recent advances in the treatment of organophosphorous poisonings. Iranian Journal of Medical Sciences, 37, 74–91.
Bigley, A. N., & Raushel, F. M. (2013). Catalytic mechanisms for phosphotriesterases. Biochimica et Biophysica Acta, 1834, 443–453.
Bisswanger, H. (2004) Enzyme reactions. In (Ed.), Practical enzymology.
Cheng, T. C. and DeFrank, J. J. (1999) Enzymatic detoxification of organophopshorus commpounds.
Cheng, T. C., Harvey, S. P., & Stroup, A. N. (1993). Purification and properties of a highly active organophosphorus acid anhydrolase from alteromonas undina. Applied and Environmental Microbiology, 59, 3138–3140.
Chernyshev, A. V., Tarasov, P. A., Semianov, K. A., Nekrasov, V. M., Hoekstra, A. G., & Maltsev, V. P. (2008). Erythrocyte lysis in isotonic solution of ammonium chloride: theoretical modeling and experimental verification. Journal of Theoretical Biology, 251, 93–107.
Colovic, M., Krstic, D., Petrovic, S., Leskovac, A., Joksic, G., Savic, J., Franko, M., Trebse, P., & Vasic, V. (2010). Toxic effects of diazinon and its photodegradation products. Toxicology Letters, 193, 9–18.
Danielsen, S., Skov, L. K., Leite, R., Laize, V. and Da Fonseca, M. L. C. (2010) DFPase enzymes from octopus vulgaris.
DeFrank, J. J., & Cheng, T. C. (1991). Purification and properties of an organophosphorus acid anhydrase from a halophilic bacterial isolate. Journal of Bacteriology, 173, 1938–1943.
Elias, M., Liebschner, D., Koepke, J., Lecomte, C., Guillot, B., Jelsch, C., & Chabriere, E. (2013). Hydrogen atoms in protein structures: high-resolution X-ray diffraction structure of the DFPase. BMC Research Notes, 6, 308.
Gab, J., Melzer, M., Kehe, K., Richardt, A., & Blum, M. M. (2009). Quantification of hydrolysis of toxic organophosphates and organophosphonates by diisopropyl fluorophosphatase from Loligo vulgaris by in situ fourier transform infrared spectroscopy. Analytical Biochemistry, 385, 187–193.
Gab, J., Melzer, M., Kehe, K., Wellert, S., Hellweg, T., & Blum, M. M. (2010). Monitoring the hydrolysis of toxic organophosphonate nerve agents in aqueous buffer and in bicontinuous microemulsions by use of diisopropyl fluorophosphatase (DFPase) with (1)H- (31)P HSQC NMR spectroscopy. Analytical and Bioanalytical Chemistry, 396, 1213–1221.
Gheybi, E., Amani, J., Salmanian, A. H., Mashayekhi, F., & Khodi, S. (2014). Designing a recombinant chimeric construct contain MUC1 and HER2 extracellular domain for prediagnostic breast cancer. Tumor Biology, 35, 11489–11497.
Gianessi, L. P. (2013). The increasing importance of herbicides in worldwide crop production. Pest Management Science, 69, 1099–1105.
Han, D., Filocamo, S., Kirby, R., & Steckl, A. (2011). Deactivating chemical agents using enzyme-coated nanofibers formed by electrospinning. ACS Applied Materials & Interfaces, 3, 4633–4639.
Hartleib, J., & Ruterjans, H. (2001). Insights into the reaction mechanism of the diisopropyl fluorophosphatase from loligo vulgaris by means of kinetic studies, chemical modification and site-directed mutagenesis. Biochimica et Biophysica Acta, 1546, 312–324.
Huang, Z., Ma, L., Liu, W., & Cheng, Y. (1999). Isotachophoresis analysis of hydrolytic products of DFP catalyzed by DFPase in porcine liver. Se Pu, 17, 196–198.
Kang, D. G., Li, L., Ha, J. H., Choi, S. S., & Cha, H. J. (2008). Efficient cell surface display of organophosphorus hydrolase using N-terminal domain of ice nucleation protein in Escherichia coli. Korean Journal of Chemistry and Engineering, 25, 804–807.
Karami, A., Latifi, A. M., & Khodi, S. (2014). Comparison of the organophosphorus hydrolase surface display using InaVN and Lpp-OmpA systems in Escherichia coli. Journal of Microbiology and Biotechnology, 24, 379–385.
Khodi, S., Latifi, A. M., Saadati, M., Mirzaei, M., & Aghamollaei, H. (2012). Surface display of organophosphorus hydrolase on E. coli using N-terminal domain of ice nucleation protein InaV. Journal of Microbiology and Biotechnology, 22, 234–238.
Kwak, Y. D., Yoo, S. K., & Kim, E. J. (1999). Cell surface display of human immunodeficiency virus type 1 gp120 on Escherichia coli by using ice nucleation protein. Clinical and Diagnostic Laboratory Immunology, 6, 499–503.
Laemmli, U. K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685.
Latifi, A. M., Khodi, S., Mirzaei, M., Miresmaeili, M., & Babavalian, H. (2012). Isolation and characterization of five chlorpyrifos degrading bacteria. African Journal of Biotechnology, 11, 3140–3146.
Lee, S. Y., Choi, J. H., & Xu, Z. H. (2003). Microbial cell-surface display. Trends in Biotechnology, 21, 45–52.
Mierendorf, R. C., Morris, B. B., Hammer, B., & Novy, R. E. (1998). Expression and purification of recombinant proteins using the pET system. Methods in Molecular Medicine, 13, 257–292.
Palaiomylitou, M. A., Kalimanis, A., Koukkou, A. I., Drainas, C., Anastassopoulos, E., Panopoulos, N. J., Ekateriniadou, L. V., & Kyriakidis, D. A. (1998). Phospholipid analysis and fractional reconstitution of the ice nucleation protein activity purified from Escherichia coli overexpressing the inaZ gene of Pseudomonas syringae. Cryobiology, 37, 67–76.
Razavi, S. M., Salamati, P., Saghafinia, M., & Abdollahi, M. (2012). A review on delayed toxic effects of sulfur mustard in Iranian veterans. Daru, 20.
Richardt, A. and Blum, M. M. (2008) Decontamination of warfare agents: enzymatic methods for the removal of B/C weapons.
Rogers, J. H. T., Kenny, D., MacGregor, I., Tracy, K., Krile, R., Nishioka, M., Taylor, M., Riggs, K., Stone, H. (2009) Decontamination of toxic industrial chemicals and chemical warfare agents on building materials using chlorine dioxide fumigant and liquid oxidant technologies.
Sarhan, M. A. A. (2011). Ice nucleation protein as a bacterial surface display protein. Archives of Biological Science Belgrade, 63, 943–948.
Sepahi, A. A., Golpasha, I. D., Emami, M., & Nakhoda, A. (2008). Isolation and characterization of crude oil degrading Bacillus spp. Iranian Journal of Environmental Health Science & Engineering, 5, 149–154.
Shimazu, M., Mulchandani, A., & Chen, W. (2001). Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnology Progress, 17, 76–80.
Singh, B. K., & Walker, A. (2006). Microbial degradation of organophosphorus compounds. FEMS Microbiology Reviews, 30, 428–471.
Stehle, R., Schulreich, C., Wellert, S., Gab, J., Blum, M. M., Kehe, K., Richardt, A., Lapp, A., & Hellweg, T. (2014). An enzyme containing microemulsion based on skin friendly oil and surfactant as decontamination medium for organo phosphates: phase behavior, structure, and enzyme activity. Journal of Colloid and Interface Science, 413, 127–132.
Tang, X., Liang, B., Yi, T., Manco, G., Palchetti, I., & Liu, A. (2014). Cell surface display of organophosphorus hydrolase for sensitive spectrophotometric detection of p-nitrophenol substituted organophosphates. Enzyme and Microbial Technology, 55, 107–112.
Theriot, C. M., & Grunden, A. M. (2011). Hydrolysis of organophosphorus compounds by microbial enzymes. Applied Microbiology and Biotechnology, 89, 35–43.
van Alphen, L., Riemens, T., Poolman, J., & Zanen, H. C. (1983). Characteristics of major outer membrane proteins of Haemophilus influenzae. Journal of Bacteriology, 155, 878–885.
Wille, T., Scott, C., Thiermann, H., & Worek, F. (2012). Detoxification of G- and V-series nerve agents by the phosphotriesterase OpdA. Biocatalysis and Biotransformation, 30, 203–208.
Wymore, T., Field, M. J., Langan, P., Smith, J. C., & Parks, J. M. (2014). Hydrolysis of DFP and the nerve agent (S)-sarin by DFPase proceeds along two different reaction pathways: implications for engineering bioscavengers. The Journal of Physical Chemistry. B, 118, 4479–4489.
Xie, J., Zhao, Y., Zhang, H., Liu, Z., & Lu, Z. (2014). Improving methyl parathion hydrolase to enhance its chlorpyrifos-hydrolysing efficiency. Letters in Applied Microbiology, 58, 53–59.
Xu, Y., Liu, Q., Zhou, L., Yang, Z., & Zhang, Y. (2008). Surface display of GFP by Pseudomonas syringae truncated ice nucleation protein in attenuated vibrio anguillarum strain. Marine Biotechnology, 10, 701–708.
Yang, C., Cai, N., Dong, M., Jiang, H., Li, J., Qiao, C., Mulchandani, A., & Chen, W. (2008). Surface display of MPH on Pseudomonas putida JS444 using ice nucleation protein and its application in detoxification of organophosphates. Biotechnology and Bioengineering, 99, 30–37.
Yang, C., Zhao, Q., Liu, Z., Li, Q., Qiao, C., Mulchandani, A., & Chen, W. (2008). Cell surface display of functional macromolecule fusions on Escherichia coli for development of an autofluorescent whole-cell biocatalyst. Environmental Science and Technology, 42, 6105–6110.
Acknowledgments
The authors would like to thank all colleagues in the Applied Biotechnology Research Center of Baqiyatallah Medical Sciences University, Analytical Chemistry and Biology departments, for their kind contribution to the research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Latifi, A.M., Karami, A. & Khodi, S. Efficient Surface Display of Diisopropylfluorophosphatase (DFPase) in E. coli for Biodegradation of Toxic Organophosphorus Compounds (DFP and Cp) . Appl Biochem Biotechnol 177, 624–636 (2015). https://doi.org/10.1007/s12010-015-1766-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-015-1766-0