Abstract
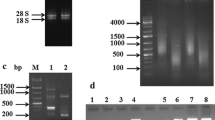
Moth bean (Vigna aconitifolia (Jacq.) Marechal), an important grain-legume crop grown in hot desert regions of Thar, under scorching sun rays, was investigated for heat tolerance at molecular level. In the present study, we constructed a forward suppression subtractive hybridization (SSH) cDNA library of heat tolerant genotype RMO-40 to identify genes expressing under delayed response to elevated temperature. Heat induction was carried out by exposing 14-day-old seedlings to elevated temperature of 42 °C for 30 min. A total of 125 unigenes (33 contigs and 92 singletons) were derived by cluster assembly and sequence alignment of 200 ESTs; out of 125 unigenes, 21 (16 %) were found to be novel to moth bean. Gene ontology functional classification terms were retrieved for 98 (78.4 %) unigenes of which 73 (58.4 %) ESTs were functionally annotated (GO consensus) where 19 unigenes were annotated with 11 enzyme commission (EC) codes and were mapped to 25 different KEGG pathways. We have identified a majority of heat-shock proteins (constituting 35 % of the present library) aiding heat stress tolerance to moth bean. An expression level of 22 ESTs generated from the above SSH cDNA library was studied through semiquantitative RT-PCR assay simultaneously under 5 and 30 min of heat stress at 42 °C.








Similar content being viewed by others
References
Baler, R., Zou, J., & Voellmy, R. (1996). Evidence for a role of Hsp70 in the regulation of the heat shock response in mammalian cells. Cell Stress Chaperones, 1, 33–39.
Callis, J., & Vierstra, R. D. (2000). Protein degradation in signaling. Current Opinion in Plant Biology, 3, 381–386.
Chen, J., Burke, J. J., Velten, J., & Xin, Z. (2006). FtsHII protease plays a critical role in Arabidopsis thermo tolerance. Plant Journal, 48(I), 73–84.
Conesa, A., Götz, S., & García-Gómez, J. M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics, 21, 3674–3676.
Craig, E. A. (1993). Chaperones: helpers along the pathways to protein folding. Science, 260, 1902–1903.
Deokar, A. A., Kondawar, V., Jain, P. K., Karuppayil, M., Raju, N. L., Vadez, V., Varshney, R. K., & Srinivasan, R. (2011). Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and susceptible genotypes of chickpea under terminal drought stress. BMC Genomics, 11, 70.
Feussner, K., Feussner, I., Leopold, I., & Wasternack, C. (1997). Isolation of a cDNA coding for an ubiquitin-conjugating enzyme UBC1 of tomato the first stress-induced UBC of higher plants. FEBS Letters, 409, 211–215.
Fink, A. K. (1999). Chaperone-mediated protein folding. Physiological Reviews, 79, 425–449.
Frydman, J., Nimmesgern, E., Ohtsuka, K., & Hartl, F. U. (1994). Folding of nascent polypeptide chains in a high molecular mass assembly with molecular chaperones. Nature, 370, 111–117.
Guisbert, E., Herman, C., Lu, C. Z., & Gross, C. A. (2004). A chaperone network controls the heat shock response in E. coli. Genes & Development, 18, 2812–2821.
Hall (2001). In: Crop response to environment. CRC press LLC, Boca Raton, Florida Pp. 232.
Hundertmark, M., & Hincha, D. K. (2008). LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics, 9, 118–139.
Koning, A. J., Rose, R., & Comai, L. (1992). Developmental expression of tomato heat-shock cognate protein 80. Plant Physiology, 100, 801–811.
Kristensen, T. N., Dahlgaard, J., & Loeschcke, V. (2002). Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evolutionary Ecology Research, 4, 1209–1216.
Kumar, R., & Singh, N. P. (1988). Effect of dates of sowing on growth and yield of cowpea (Vignaunguiculata (L.) Walp). Legume Research, 21(1), 54–56.
Mario, H., Mahadi, B., Francois, O., Jean, D., Antonio, F. M., Ani, D., Patrick, G., Anne, B., Andre, L., Mathew, G. L., Luke, M., William, L. C., Fathey, S. (2006). Wheat EST resource for functional genomics of abiotic stress. BMC Genomics 7-149.
Mullarkey, M., & Jones, P. (2000). Isolation and analysis of thermotolerant mutant in wheat. Journal of Experimental Botany, 51(342), 139–146.
Nishikawa, S., & Toshiya, E. (1997). The yeast JEM1p is a DnaJ-like protein of the endoplasmic reticulum membrane required for nuclear fusion. Journal of Biological Chemistry, 16, 12889–12892.
Noh, S. J., Kwon, C. S., Oh, D. H., Moon, J. S., & Chung, W. I. (2003). Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene, 311, 81–91.
Okuda, S., Yamada, T., Hamajima, M., Itoh, M., Katayama, T., Bork, P., Goto, S., & Kanehisa, M. (2008). KEGG atlas mapping for global analysis of metabolic pathways. Nucleic acid research, 36, W423–W426.
Prodromou, C., Roe, S. M., Piper, P. W., & Pearl, L. H. (1997). A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Natural Structural Biology, 4, 477–482.
Rampuria, S., Joshi, U., Palit, P., Deokar, A. A., Meghwal, R. R., Mohapatra, T., Srinivasan, R., Bhatt, K. V., & Sharma, R. (2012). Construction and analysis of an SSH cDNA library of early heat induced genes of Vigna aconitifolia variety RMO-40. Genome, 55, 783–796.
Sakaki, K., Tashiro, K., Kuhara, S., & Mihara, K. (2003). Response of genes associated with mitochondrial function to mild heat stress in yeast Saccharomyces cerevisiae. Journal of Biochemistry, 134, 373–384.
Schoffl, F., Prandl, R., & Reindl, A. (1998). Regulation of the heat-shock response. Plant Physiology, 117(4), 1135–1141.
Sharma, R., Jain, M., Kumar, S., & Kumar, V. (2014). Evaluation of differences among Vignaaconitifolia varieties for acquired thermotolerance. Agricultural Research. doi:10.1007/s40003-014-0108-8.
Sung, D. Y., Vierling, E., & Guy, C. L. (2001). Comprehensive expression profile analysis of the Arabidopsis Hsp70 Gene Family1. Plant Physiology, 126(2), 789–800.
Tian, J., Belangera, C. F., & Huang, B. (2009). Identification of heat stress-responsive genes in heat-adapted thermal Agrostis scabra by suppression subtractive hybridization. Journal of Plant Physiology, 166, 588–601.
Wahid, A. (2007). Physiological implications of metabolites biosynthesis in net assimilation and heat stress tolerance of sugarcane sprouts. J. Plants Research, 120, 219–228.
Wheeler, J. C., King, V., & Tower, J. (1999). Sequence requirements for upregulating expression of Drosophila hsp70 transgenes during aging. Neurobiology of Aging, 20, 545–553.
Zhang, Y., Mian, M. A. R., Chekhovskiy, K., So, S., Kupfer, D., Lai, H., & Roe, B. A. (2005). Differential gene expression in Festuca under heat stress conditions. Journal of Experimental Botany, 56, 897–907.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gurjar, K., Rampuria, S., Joshi, U. et al. Identification of Heat-Related ESTs in Moth Bean Through Suppression Subtraction Hybridization. Appl Biochem Biotechnol 173, 2116–2128 (2014). https://doi.org/10.1007/s12010-014-1011-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-014-1011-2