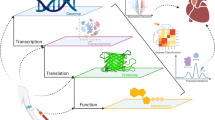
Abstract
Purpose of review
Cardiac imaging and sequencing have greatly improved over the recent years. The goal of this review is to summarize these recent advances in cardiac imaging and sequencing, their application in heart-transplantation, and provide our perspective in how artificial intelligence provides a new paradigm for big data-driven analysis in heart-transplant research.
Recent findings
Cardiac imaging, particularly parametric mapping by cardiac MRI and global longitudinal strain by echocardiography, has improved our understanding of cardiac allograft rejection and prediction of adverse clinical outcomes. Independently, gene expression profiling and measurement of donor-derived cell-free DNA have greatly improved risk stratification for acute rejection. More recently, data-driven phenotypic clustering using novel machine learning algorithms has been used to identify a distinct macrophage subset, associated with acute rejection.
Summary
Developments in imaging and sequencing techniques in the application of heart-transplant research are improving rapidly and in parallel with improvements in analysis of these large datasets. The approach to heart-transplant research is in the transition of significant change as big data-driven analysis identifies new mechanistic patterns that can be combined with traditional hypothesis testing.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
McMurray JJV, Solomon SD, Inzucchi SE, et al. Dapagliflozin in patients with heart failure and reduced ejection fraction. N Engl J Med. 2019;381:1995–2008.
McMurray JJV, Packer M, Desai AS, Gong J, Lefkowitz MP, Rizkala AR, et al. Angiotensin-neprilysin inhibition versus enalapril in heart failure. N Engl J Med. 2014;371:993–1004.
Shen L, Jhund PS, McMurray JJV. Declining risk of sudden death in heart failure. N Engl J Med. 2017;377:1794–5.
Kittleson MM. New issues in heart transplantation for heart failure. Curr Treat Options Cardiovasc Med. 2012;14:356–69.
Khush KK, Cherikh WS, Chambers DC, Harhay MO, Hayes D Jr, Hsich E, et al. The International Thoracic Organ Transplant Registry of the International Society for Heart and Lung Transplantation: thirty-sixth adult heart transplantation report - 2019; focus theme: donor and recipient size match. J Heart Lung Transplant. 2019;38:1056–66.
Costanzo MR, Dipchand A, Starling R, et al. The International Society of Heart and Lung Transplantation Guidelines for the care of heart transplant recipients. J Heart Lung Transplant. 2010;29:914–56.
Mathier MA, McNamara DM. Management of the patient after heart transplant. Curr Treat Options Cardiovasc Med. 2004;6:459–69.
Olymbios M, Kwiecinski J, Berman DS, Kobashigawa JA. Imaging in heart transplant patients. JACC Cardiovasc Imaging. 2018;11:1514–30.
•• Fernandez DM, Rahman AH, Fernandez NF, et al. Single-cell immune landscape of human atherosclerotic plaques. Nat Med. 2019;25:1576–88 Landmark study demonstrating the use of cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) for single cell proteomic and transcriptomic analysis in atherosclerosis to determine a distinct subset of activated CD4+ T cells.
Mingo-Santos S, Moñivas-Palomero V, Garcia-Lunar I, Mitroi CD, Goirigolzarri-Artaza J, Rivero B, et al. Usefulness of two-dimensional strain parameters to diagnose acute rejection after heart transplantation. J Am Soc Echocardiogr. 2015;28:1149–56.
• Butler CR, Savu A, Bakal JA, et al. Correlation of cardiovascular magnetic resonance imaging findings and endomyocardial biopsy results in patients undergoing screening for heart transplant rejection. J Heart Lung Transplant. 2015;34:643–50 This prospective study showed the use of T2 relaxation time and right ventricular end-diastolic volume index by CMR that predicted a positive endomyocardial biopsy with good accuracy.
Dolan RS, Rahsepar AA, Blaisdell J, Suwa K, Ghafourian K, Wilcox JE, et al. Multiparametric cardiac magnetic resonance imaging can detect acute cardiac allograft rejection after heart transplantation. JACC Cardiovasc Imaging. 2019;12:1632–41.
Vermes E, Pantaléon C, Auvet A, Cazeneuve N, Machet MC, Delhommais A, et al. Cardiovascular magnetic resonance in heart transplant patients: diagnostic value of quantitative tissue markers: T2 mapping and extracellular volume fraction, for acute rejection diagnosis. J Cardiovasc Magn Reson. 2018;20:59.
Akosah KO, McDaniel S, Hanrahan JS, Mohanty PK. Dobutamine stress echocardiography early after heart transplantation predicts development of allograft coronary artery disease and outcome. J Am Coll Cardiol. 1998;31:1607–14.
Chirakarnjanakorn S, Starling RC, Popović ZB, Griffin BP, Desai MY. Dobutamine stress echocardiography during follow-up surveillance in heart transplant patients: diagnostic accuracy and predictors of outcomes. J Heart Lung Transplant. 2015;34:710–7.
• Miller CA, Sarma J, Naish JH, et al. Multiparametric cardiovascular magnetic resonance assessment of cardiac allograft vasculopathy. J Am Coll Cardiol. 2014;63:799–808. This is one of the earliest studies demonstrating the high sensitivity of stress perfusion CMR to detect both macro- and microvascular CAV.
• Braggion-Santos MF, Lossnitzer D, Buss S, Lehrke S, Doesch A, Giannitsis E, et al. Late gadolinium enhancement assessed by cardiac magnetic resonance imaging in heart transplant recipients with different stages of cardiac allograft vasculopathy. Eur Heart J Cardiovasc Imaging. 2014;15:1125–32 This study details LGE distribution across different ISHLT CAV grades. Shows both infarct-typical and infarct-atypical patterns in more severe forms of CAV.
•• Chih S, Ross HJ, Alba AC, Fan CS, Manlhiot C, Crean AM. Perfusion cardiac magnetic resonance imaging as a rule-out test for cardiac allograft vasculopathy. Am J Transplant. 2016;16:3007–15 This prospective, cross-sectional study showed stress perfusion CMR is a sensitive noninvasive test for CAV compared to IVUS using a myocardial perfusion reserve cut-off of 1.68.
Erbel C, Mukhammadaminova N, Gleissner CA, Osman NF, Hofmann NP, Steuer C, et al. Myocardial perfusion reserve and strain-encoded CMR for evaluation of cardiac allograft microvasculopathy. JACC Cardiovasc Imaging. 2016;9:255–66.
Miller RJH, Kwiecinski J, Shah KS, Eisenberg E, Patel J, Kobashigawa JA, et al. Coronary computed tomography-angiography quantitative plaque analysis improves detection of early cardiac allograft vasculopathy: a pilot study. Am J Transplant. 2020;20:1375–83.
Wever-Pinzon O, Romero J, Kelesidis I, et al. Coronary computed tomography angiography for the detection of cardiac allograft vasculopathy: a meta-analysis of prospective trials. J Am Coll Cardiol. 2014;63:1992–2004.
• Chih S, Chong AY, Erthal F, et al. PET assessment of epicardial intimal disease and microvascular dysfunction in cardiac allograft vasculopathy. J Am Coll Cardiol. 2018;71:1444–56 This study demonstrated PET myocardial flow quantification with rubidium 82 sensitively detected epicardial CAV when compared to IVUS.
Bravo PE, Bergmark BA, Vita T, Taqueti VR, Gupta A, Seidelmann S, et al. Diagnostic and prognostic value of myocardial blood flow quantification as non-invasive indicator of cardiac allograft vasculopathy. Eur Heart J. 2018;39:316–23.
Chih S, Chong AY, Bernick J, Wells GA, deKemp RA, Davies RA, et al. Validation of multiparametric rubidium-82 PET myocardial blood flow quantification for cardiac allograft vasculopathy surveillance. J Nucl Cardiol. 2020. https://doi.org/10.1007/s12350-020-02038-y.
Dandel M, Hetzer R. Post-transplant surveillance for acute rejection and allograft vasculopathy by echocardiography: usefulness of myocardial velocity and deformation imaging. J Heart Lung Transplant. 2017;36:117–31.
Kobayashi Y, Sudini NL, Rhee J-W, et al. Incremental value of deformation imaging and hemodynamics following heart transplantation: insights from graft function profiling. JACC Heart Fail. 2017;5:930–9.
Clemmensen TS, Eiskjær H, Løgstrup BB, Ilkjær LB, Poulsen SH. Left ventricular global longitudinal strain predicts major adverse cardiac events and all-cause mortality in heart transplant patients. J Heart Lung Transplant. 2017;36:567–76.
Butler CR, Kim DH, Chow K, Toma M, Thompson R, Mengel M, et al. Cardiovascular MRI predicts 5-year adverse clinical outcome in heart transplant recipients: cardiovascular MRI predicts heart transplant outcome. Am J Transplant. 2014;14:2055–61.
Pedrotti P, Vittori C, Facchetti R, Pedretti S, Dellegrottaglie S, Milazzo A, et al. Prognostic impact of late gadolinium enhancement in the risk stratification of heart transplant patients. Eur Heart J Cardiovasc Imaging. 2017;18:130–7.
Chaikriangkrai K, Abbasi MA, Sarnari R, et al. Prognostic value of myocardial extracellular volume fraction and T2-mapping in heart transplant patients. JACC Cardiovasc Imaging. 2020. https://doi.org/10.1016/j.jcmg.2020.01.014 This study showed the utility of T2 relaxation time and ECV by CMR for predicting major adverse cardiovascular events in heart-transplant patients with a median follow-up of 2.4 to 3.5 years.
Andrew H, Osama O, Afshin F-F, Felipe K, Nijjar PS, Pratik V, et al. Myocardial fibrosis and prognosis in heart transplant recipients. Circ Cardiovasc Imaging. 2019;12:e009060.
Kazmirczak F, Nijjar PS, Zhang L, Hughes A, Chen K-HA, Okasha O, et al. Safety and prognostic value of regadenoson stress cardiovascular magnetic resonance imaging in heart transplant recipients. J Cardiovasc Magn Reson. 2019;21:9.
Rohnean A, Houyel L, Sigal-Cinqualbre A, To N-T, Elfassy E, Paul J-F. Heart transplant patient outcomes: 5-year mean follow-up by coronary computed tomography angiography. Transplantation. 2011;91:583–8.
Okada K, Kitahara H, Yang H-M, Tanaka S, Kobayashi Y, Kimura T, et al. Paradoxical vessel remodeling of the proximal segment of the left anterior descending artery predicts long-term mortality after heart transplantation. JACC Heart Fail. 2015;3:942–52.
Konerman MC, Lazarus JJ, Weinberg RL, et al. Reduced myocardial flow reserve by positron emission tomography predicts cardiovascular events after cardiac transplantation. Circ Heart Fail. 2018;11:e004473.
Mc Ardle Brian A, Davies RA, Li C, et al. Prognostic value of Rubidium-82 positron emission tomography in patients after heart transplant. Circ Cardiovasc Imaging. 2014;7:930–7.
Feher A, Srivastava A, Quail MA, Boutagy NE, Khanna P, Wilson L, et al. Serial assessment of coronary flow reserve by Rubidium-82 positron emission tomography predicts mortality in heart transplant recipients. JACC Cardiovasc Imaging. 2020;13:109–20.
•• Pham MX, Teuteberg JJ, Kfoury AG, et al. Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med. 2010;362:1890–900 This is the pivotal IMAGE trial that showed noninferiority of AlloMap to surveillance endomyocardial biopsies with similar clinical outcomes in both approaches.
Lu W, Zheng J, Pan X-D, Zhang M-D, Zhu T-Y, Li B, et al. Diagnostic performance of cardiac magnetic resonance for the detection of acute cardiac allograft rejection: a systematic review and meta-analysis. J Thorac Dis. 2015;7:252–63.
Coelho-Filho OR, Shah R, Lavagnoli CFR, et al. Myocardial tissue remodeling after orthotopic heart transplantation: a pilot cardiac magnetic resonance study. Int J Card Imaging. 2018;34:15–24.
Tang Z, Kobashigawa J, Rafiei M, Stern LK, Hamilton M. The natural history of biopsy-negative rejection after heart transplantation. J Transp Secur. 2013;2013:236720.
Pedrotti P, Bonacina E, Vittori C, Frigerio M, Roghi A. Pathologic correlates of late gadolinium enhancement cardiovascular magnetic resonance in a heart transplant patient. Cardiovasc Pathol. 2015;24:247–9.
Miller RJH, Thomson L, Levine R, Dimbil SJ, Patel J, Kobashigawa JA, et al. Quantitative myocardial tissue characterization by cardiac magnetic resonance in heart transplant patients with suspected cardiac rejection. Clin Transpl. 2019;33:e13704.
Vajapey R, Eck B, Tang W, Kwon DH. Advances in MRI applications to diagnose and manage cardiomyopathies. Curr Treat Options Cardiovasc Med. 2019;21:74.
• Chaikriangkrai K, Abbasi MA, Sarnari R, et al. Natural history of myocardial late gadolinium enhancement predicts adverse clinical events in heart transplant recipients. JACC Cardiovasc Imaging. 2019;12:2092–4 This study showed that heart-transplant patients had a high prevalence of LGE that generally stayed stable during longitudinal follow-up with repeat CMR studies. Higher burden of LGE was associated with major adverse cardiovascular events.
Yuan Y, Cai J, Cui Y, Wang J, Alwalid O, Shen X, et al. CMR-derived extracellular volume fraction (ECV) in asymptomatic heart transplant recipients: correlations with clinical features and myocardial edema. Int J Card Imaging. 2018;34:1959–67.
Sade LE, Hazirolan T, Kozan H, Ozdemir H, Hayran M, Eroglu S, et al. T1 mapping by cardiac magnetic resonance and multidimensional speckle-tracking strain by echocardiography for the detection of acute cellular rejection in cardiac allograft recipients. JACC Cardiovasc Imaging. 2019;12:1601–14.
Chen Y, Zhang L, Liu J, Zhang P, Chen X, Xie M. Molecular imaging of acute cardiac transplant rejection: animal experiments and prospects. Transplantation. 2017;101:1977–86.
Stendardi W, Kim P, Hsiao A. Molecular imaging of the transplanted heart: a mechanistic approach to graft survival. Curr Cardiovasc Imaging Rep. 2017. https://doi.org/10.1007/s12410-017-9422-4.
Chih S, Chong AY, Mielniczuk LM, Bhatt DL, Beanlands RSB. Allograft vasculopathy: the Achilles’ heel of heart transplantation. J Am Coll Cardiol. 2016;68:80–91.
Mehra MR, Crespo-Leiro MG, Dipchand A, Ensminger SM, Hiemann NE, Kobashigawa JA, et al. International Society for Heart and Lung Transplantation working formulation of a standardized nomenclature for cardiac allograft vasculopathy-2010. J Heart Lung Transplant. 2010;29:717–27.
Chen MH, Abernathey E, Lunze F, Colan SD, O’Neill S, Bergersen L, et al. Utility of exercise stress echocardiography in pediatric cardiac transplant recipients: a single-center experience. J Heart Lung Transplant. 2012;31:517–23.
Clemmensen TS, Løgstrup BB, Eiskjær H, Poulsen SH. Evaluation of longitudinal myocardial deformation by 2-dimensional speckle-tracking echocardiography in heart transplant recipients: relation to coronary allograft vasculopathy. J Heart Lung Transplant. 2015;34:195–203.
Elkaryoni A, Abu-Sheasha G, Altibi AM, Hassan A, Ellakany K, Nanda NC. Diagnostic accuracy of dobutamine stress echocardiography in the detection of cardiac allograft vasculopathy in heart transplant recipients: a systematic review and meta-analysis study. Echocardiography. 2019;36:528–36.
Clerkin KJ, Farr MA, Restaino SW, Ali ZA, Mancini DM. Dobutamine stress echocardiography is inadequate to detect early cardiac allograft vasculopathy. J Heart Lung Transplant. 2016;35:1040–1.
Stocker TJ, Leipsic J, Hadamitzky M, Chen MY, Rubinshtein R, Deseive S, et al. Application of low tube potentials in CCTA: results from the PROTECTION VI study. JACC Cardiovasc Imaging. 2020;13:425–34.
von Ziegler F, Rümmler J, Kaczmarek I, Greif M, Schenzle J, Helbig S, et al. Detection of significant coronary artery stenosis with cardiac dual-source computed tomography angiography in heart transplant recipients: coronary DSCT angiography in HTX patients. Transpl Int. 2012;25:1065–71.
• Asleh R, Briasoulis A, Kremers WK, et al. Long-term sirolimus for primary immunosuppression in heart transplant recipients. J Am Coll Cardiol. 2018;71:636–50 This article describes a single-center, retrospective study of 402 heart-transplant patients and shows an association with early conversion to sirolimus from a calcineurin inhibitor with improved survival and decreased CAV-related events.
Ciliberto G. Resting echocardiography and quantitative dipyridamole technetium-99m sestamibi tomography in the identification of cardiac allograft vasculopathy and the prediction of long-term prognosis after heart transplantation. Eur Heart J. 2001;22:964–71.
Badano LP, Miglioranza MH, Edvardsen T, Colafranceschi AS, Muraru D, Bacal F, et al. European Association of Cardiovascular Imaging/Cardiovascular Imaging Department of the Brazilian Society of Cardiology recommendations for the use of cardiac imaging to assess and follow patients after heart transplantation. Eur Heart J Cardiovasc Imaging. 2015;16:919–48.
Cavalcante JL, Barboza J, Ananthasubramaniam K. Regadenoson is a safe and well-tolerated pharmacological stress agent for myocardial perfusion imaging in post-heart transplant patients. J Nucl Cardiol. 2011;18:628–33.
Manrique A, Bernard M, Hitzel A, Bubenheim M, Tron C, Agostini D, et al. Diagnostic and prognostic value of myocardial perfusion gated SPECT in orthotopic heart transplant recipients. J Nucl Cardiol. 2010;17:197–206.
Miller RJH, Kobashigawa JA, Berman DS. Should positron emission tomography be the standard of care for non-invasive surveillance following cardiac transplantation? J Nucl Cardiol. 2019;26:655–9.
Hlatky MA, Shilane D, Hachamovitch R, Dicarli MF, Investigators SPARC. Economic outcomes in the study of myocardial perfusion and coronary anatomy imaging roles in coronary artery disease registry: the SPARC study. J Am Coll Cardiol. 2014;63:1002–8.
Steen H, Merten C, Refle S, Klingenberg R, Dengler T, Giannitsis E, et al. Prevalence of different gadolinium enhancement patterns in patients after heart transplantation. J Am Coll Cardiol. 2008;52:1160–7.
Almufleh A, Garuba H, Mielniczuk LM, Davies RA, Stadnick E, Belanger E, et al. Diffuse subepicardial late gadolinium enhancement after heart transplant: a potentially ominous finding. Can J Cardiol. 2018;34:1687.e3–7.
Narang A, Blair JE, Patel MB, Mor-Avi V, Fedson SE, Uriel N, et al. Myocardial perfusion reserve and global longitudinal strain as potential markers of coronary allograft vasculopathy in late-stage orthotopic heart transplantation. Int J Card Imaging. 2018;34:1607–17.
Yang H-M, Khush K, Luikart H, Okada K, Lim H-S, Kobayashi Y, et al. Invasive assessment of coronary physiology predicts late mortality after heart transplantation. Circulation. 2016;133:1945–50.
•• Hsu L-Y, Jacobs M, Benovoy M, et al. Diagnostic performance of fully automated pixel-wise quantitative myocardial perfusion imaging by cardiovascular magnetic resonance. JACC Cardiovasc Imaging. 2018;11:697–707 One of the earliest studies to show the accuracy of automatic motion correction and quantification of absolute myocardial blood flow by CMR, paving the way for its clinical use in patients with coronary artery disease.
Knott KD, Seraphim A, Augusto JB, et al. The prognostic significance of quantitative myocardial Perfusion: An artificial intelligence based approach using perfusion mapping. Circulation. 2020. https://doi.org/10.1161/circulationaha.119.044666.
Clemmensen TS, Eiskjaer H, Løgstrup BB, Valen KPB, Mellemkjaer S, Poulsen SH. Prognostic value of exercise myocardial deformation and haemodynamics in long-term heart-transplanted patients. ESC Heart Fail. 2019;6:629–39.
Ide S, Riesenkampff E, Chiasson DA, Dipchand AI, Kantor PF, Chaturvedi RR, et al. Histological validation of cardiovascular magnetic resonance T1 mapping markers of myocardial fibrosis in paediatric heart transplant recipients. J Cardiovasc Magn Reson. 2017;19:10.
Shenoy C, Romano S, Hughes A, Okasha O, Nijjar PS, Velangi P, et al. Cardiac magnetic resonance feature tracking global longitudinal strain and prognosis after heart transplantation. J Am Coll Cardiol Img. 2020. https://doi.org/10.1016/j.jcmg.2020.04.004.
Ryan CM, Chaudhuri A, Concepcion W, Grimm PC. Immune cell function assay does not identify biopsy-proven pediatric renal allograft rejection or infection. Pediatr Transplant. 2014;18:446–52.
Ling X, Xiong J, Liang W, Schroder PM, Wu L, Ju W, et al. Can immune cell function assay identify patients at risk of infection or rejection? A meta-analysis. Transplantation. 2012;93:737–43.
Deng MC. The AlloMap™ genomic biomarker story: 10 years after. Clin Transpl. 2017. https://doi.org/10.1111/ctr.12900 A review article describing the purpose, development and implementation of gene expression profiling using AlloMap in heart-transplantation.
Crespo-Leiro MG, Stypmann J, Schulz U, et al. Clinical usefulness of gene-expression profile to rule out acute rejection after heart transplantation: CARGO II. Eur Heart J. 2016;37:2591–601.
Colvin MM, Cook JL, Chang P, et al. Antibody-mediated rejection in cardiac transplantation: emerging knowledge in diagnosis and management. Circulation. 2015;131:1608–39.
• Wehmeier C, Hönger G, Cun H, Amico P, Hirt-Minkowski P, Georgalis A, et al. Donor specificity but not broadness of sensitization is associated with antibody-mediated rejection and graft loss in renal allograft recipients. Am J Transplant. 2017;17:2092–102 This study showed that donor specific antibody profile was more predictive of antibody-mediated rejection and allograft survival than pre-kidney transplant calculated panel of reactive antibodies score.
Butler CL, Valenzuela NM, Thomas KA, Reed EF. Not all antibodies are created equal: factors that influence antibody mediated rejection. J Immunol Res. 2017;2017:7903471.
•• Khush KK, Patel J, Pinney S, et al. Noninvasive detection of graft injury after heart transplant using donor-derived cell-free DNA: A prospective multicenter study. Am J Transplant. 2019;19:2889–99 This study established the use of dd-cfDNA to detect ACR and AMR in heart-transplant patients with a cut-off of 0.2%.
•• Bloom RD, Bromberg JS, Poggio ED, et al. Cell-free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017;28:2221–32 DART study demonstrating that dd-cfDNA can be used to detect kidney allograft injury from T cell- and antibody-mediated rejection using a 1% cut-off.
Loupy A, Duong Van Huyen JP, Hidalgo L, et al. Gene expression profiling for the identification and classification of antibody-mediated heart rejection. Circulation. 2017;135:917–35.
•• Liu P, Tseng G, Wang Z, Huang Y, Randhawa P. Diagnosis of T-cell–mediated kidney rejection in formalin-fixed, paraffin-embedded tissues using RNA-Seq–based machine learning algorithms. Hum Pathol. 2019;84:283–90 This study showed the application of machine learning on bulk RNA-Seq data from formalin-fixed, paraffin-embedded kidney biopsy tissue to determine T cell-mediated rejection in kidney-transplant patients. Additionally, the authors showed the high prevalence of mixed rejection--both T cell- and antibody-mediated rejection.
Reeve J, Böhmig GA, Eskandary F, Einecke G, Lefaucheur C, Loupy A, et al. Assessing rejection-related disease in kidney transplant biopsies based on archetypal analysis of molecular phenotypes. JCI insight. 2017. https://doi.org/10.1172/jci.insight.94197 This study showed the use of machine learning with array-based data from kidney-transplant biopsies for creation of six archetypes, with classifications beyond standard histopathology.
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7.
•• Newman AM, Steen CB, Liu CL, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat Biotechnol. 2019;37:773–82 Landmark study demonstrating the application of CIBERSORTx for deconvolution of different cell types without need for physical cell isolation.
McArdle S, Buscher K, Ehinger E, Pramod AB, Riley N, Ley K (2018) PRESTO, a new tool for integrating large-scale -omics data and discovering disease-specific signatures. bioRxiv 302604. This paper describes the novel machine learning technique of PREdictive Stochastic neighbor embedding Tool for Omics’ or PRESTO for unsupervised dimensionality reduction of multivariate data matrices, such as microarray and proteomic datasets.
Higdon LE, Schaffert S, Khatri P, Maltzman JS. Single cell immune profiling in transplantation research. Am J Transplant. 2019;19:1278–87.
•• Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. 2018;29:2069–80 This paper describes the application of single cell RNA-Seq in a kidney-transplant patient, showing two distinct monocyte clusters as well as two distinct pathologic endothelial cell responses.
Winkels H, Ehinger E, Vassallo M, Buscher K, Dinh HQ, Kobiyama K, et al. Atlas of the immune cell repertoire in mouse atherosclerosis defined by single-cell RNA-sequencing and mass cytometry. Circ Res. 2018;122:1675–88.
Kowalski R, Post D, Schneider MC, Britz J, Thomas J, Deierhoi M, et al. Immune cell function testing: an adjunct to therapeutic drug monitoring in transplant patient management. Clin Transpl. 2003;17:77–88.
Kobashigawa JA, Kiyosaki KK, Patel JK, Kittleson MM, Kubak BM, Davis SN, et al. Benefit of immune monitoring in heart transplant patients using ATP production in activated lymphocytes. J Heart Lung Transplant. 2010;29:504–8.
Libri I, Gnappi E, Zanelli P, Reina M, Giuliodori S, Vaglio A, et al. Trends in immune cell function assay and donor-specific HLA antibodies in kidney transplantation: a 3-year prospective study. Am J Transplant. 2013;13:3215–22.
Mehrotra A, Leventhal J, Purroy C, Cravedi P. Monitoring T cell alloreactivity. Transplant Rev. 2015;29:53–9.
Askar M. T helper subsets & regulatory T cells: rethinking the paradigm in the clinical context of solid organ transplantation. Int J Immunogenet. 2014;41:185–94.
Jon K, Jignesh P, Babak A, et al. Randomized pilot trial of gene expression profiling versus heart biopsy in the first year after heart transplant. Circ Heart Fail. 2015;8:557–64.
Moayedi Y, Foroutan F, Miller RJH, Fan CPS, Posada JGD, Alhussein M, et al. Risk evaluation using gene expression screening to monitor for acute cellular rejection in heart transplant recipients. J Heart Lung Transplant. 2019;38:51–8.
De Vlaminck I, Valantine HA, Snyder TM, et al. Circulating cell-free DNA enables noninvasive diagnosis of heart transplant rejection. Sci Transl Med. 2014;6:241ra77.
Ying L, Sarwal M. In praise of arrays. Pediatr Nephrol. 2009;24:1643–59 quiz 1655, 1659.
Reeve J, Sellarés J, Mengel M, Sis B, Skene A, Hidalgo L, et al. Molecular diagnosis of T cell-mediated rejection in human kidney transplant biopsies. Am J Transplant. 2013;13:645–55.
Halloran PF, Potena L, Van Huyen J-PD, Bruneval P, Leone O, Kim DH, et al. Building a tissue-based molecular diagnostic system in heart transplant rejection: the heart molecular microscope diagnostic (MMDx) system. J Heart Lung Transplant. 2017;36:1192–200.
• Parkes MD, Aliabadi AZ, Cadeiras M, et al. An integrated molecular diagnostic report for heart transplant biopsies using an ensemble of diagnostic algorithms. J Heart Lung Transplant. 2019;38:636–46 This study demonstrated the use of machine learning to identify molecular archetypes from array-based endomyocardial biopsy tissue data, with more accurate performance compared to histopathological diagnosis.
Chartrand G, Cheng PM, Vorontsov E, Drozdzal M, Turcotte S, Pal CJ, et al. Deep learning: a primer for radiologists. Radiographics. 2017;37:2113–31.
Cochain C, Vafadarnejad E, Arampatzi P, Pelisek J, Winkels H, Ley K, et al. Single-cell RNA-Seq reveals the transcriptional landscape and heterogeneity of aortic macrophages in murine atherosclerosis. Circ Res. 2018;122:1661–74.
Seetharam K, Kagiyama N, Shrestha S, Sengupta PP. Clinical inference from cardiovascular imaging: paradigm shift towards machine-based intelligent platform. Curr Treat Options Cardiovasc Med. 2020;22:8.
Krittanawong C, Johnson KW, Tang WW. How artificial intelligence could redefine clinical trials in cardiovascular medicine: lessons learned from oncology. Perinat Med. 2019;16:83–8.
Haro Alonso D, Wernick MN, Yang Y, Germano G, Berman DS, Slomka P. Prediction of cardiac death after adenosine myocardial perfusion SPECT based on machine learning. J Nucl Cardiol. 2019;26:1746–54.
Schlesinger DE, Stultz CM. Deep learning for cardiovascular risk stratification. Curr Treat Options Cardiovasc Med. 2020;22:15.
Retson TA, Masutani EM, Golden D, Hsiao A. Clinical performance and role of expert supervision of deep learning for cardiac ventricular volumetry: a validation study. Radiology: Artificial Intelligence. 2020;2:e190064.
Blansit K, Retson T, Masutani E, Bahrami N, Hsiao A. Deep learning-based prescription of cardiac MRI planes. Radiol Artif Intell. 2019;1:e180069.
Masutani EM, Bahrami N, Hsiao A. Deep learning single-frame and multiframe super-resolution for cardiac MRI. Radiology. 2020;295:552–61.
McCann MT, Jin KH, Unser M. Convolutional neural networks for inverse problems in imaging: a review. IEEE Signal Process Mag. 2017;34:85–95.
Gulati A, Ismail TF, Ali A, Hsu LY, Gonçalves C, Ismail NA, et al. Microvascular dysfunction in dilated cardiomyopathy. J Am Coll Cardiol Img. 2019;12:1699–708.
Arai AE, Hsu L-Y. Global developments in stress perfusion cardiovascular magnetic resonance. Circulation. 2020;141:1292–4.
Lin DJ, Johnson PM, Knoll F, Lui YW. Artificial intelligence for MR image reconstruction: an overview for clinicians. J Magn Reson Imaging. 2020. https://doi.org/10.1002/jmri.27078.
Nirschl JJ, Janowczyk A, Peyster EG, Frank R, Margulies KB, Feldman MD, et al. Deep learning tissue segmentation in cardiac histopathology images. Deep Learn Med Image Anal. 2017:179–95.
Litjens G, Sánchez CI, Timofeeva N, Hermsen M, Nagtegaal I, Kovacs I, et al. Deep learning as a tool for increased accuracy and efficiency of histopathological diagnosis. Sci Rep. 2016. https://doi.org/10.1038/srep26286.
Becker JU, Mayerich D, Padmanabhan M, Barratt J, Ernst A, Boor P, et al. Artificial intelligence and machine learning in nephropathology. Kidney Int. 2020;98:65–75.
Peyster EG, Madabhushi A, Margulies KB. Advanced morphologic analysis for diagnosing allograft rejection: the case of cardiac transplant rejection. Transplantation. 2018;102:1230–9.
Halloran PF, Reeve J, Aliabadi AZ, Cadeiras M, Crespo-Leiro MG, Deng M, et al. Exploring the cardiac response to injury in heart transplant biopsies. JCI Insight. 2018;3. https://doi.org/10.1172/jci.insight.123674.
•• Loupy A, Aubert O, Orandi BJ, et al. Prediction system for risk of allograft loss in patients receiving kidney transplants: international derivation and validation study. BMJ. 2019;366:l4923 This study created the iBox system for prediction of allograft loss in kidney-transplant patients using eight functional, histological and immunological prognostic factors.
•• Loupy A, Coutance G, Bonnet G, et al. Identification and characterization of trajectories of cardiac allograft vasculopathy after heart transplantation: a population-based study. Circulation. 2020;141:1954–67 This study identified six independent factors (donor age, donor sex, donor tobacco use, recipient dyslipidemia, class II donor specific antibodies, ACR episode) to predict CAV trajectory using unsupervised latent class mixed models.
van Assen M, Cornelissen LJ. Artificial Intelligence. J Am Coll Cardiol Img. 2020;13:1172–4.
Litjens G, Ciompi F, Wolterink JM, de Vos BD, Leiner T, Teuwen J, et al. State-of-the-art deep learning in cardiovascular image analysis. JACC Cardiovasc Imaging. 2019;12:1549–65.
Chong AS, Alegre M-L, Miller ML, Fairchild RL. Lessons and limits of mouse models. Cold Spring Harb Perspect Med. 2013;3:a015495.
Funding
This work was supported by the American Heart Association Career Development Grant #18CDA34110250/Paul J. Kim, MD/2018.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr. Paul J. Kim reports grants from CareDx, grants from Astellas Pharma,outside the submitted work.
Carol E. Battikha and Ibrahim Selevany declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Heart Failure
Rights and permissions
About this article
Cite this article
Battikha, C.E., Selevany, I. & Kim, P.J. Advances and New Insights in Post-Transplant Care: From Sequencing to Imaging. Curr Treat Options Cardio Med 22, 32 (2020). https://doi.org/10.1007/s11936-020-00828-8
Published:
DOI: https://doi.org/10.1007/s11936-020-00828-8