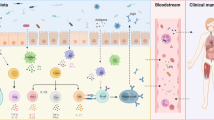
Abstract
Purpose of Review
Systemic lupus erythematosus (SLE) is a prototypic autoimmune disease that typically displays chronic inflammatory tissue damage and miscellaneous circulatory autoantibodies, as well as distinctive type 1 interferon signatures. The etiology of SLE is unclear and currently is attributed to genetic predisposition and environmental triggers. Gut microbiota has recently been considered a critical environmental pathogenic factor in immune-related disorders, and studies are ongoing to uncover the key pathogens and the imputative mechanisms. Fundamental advancements on the role of the microbiota in SLE pathology have been achieved in recent years and are summarized in this review.
Recent Findings
Recent findings suggested that gut commensals could propagate autoimmunity via molecular mimicry in which ortholog-carrying microbes cross-activate autoreactive T/B cells and trigger the response against host autoantigens, or via bystander activation by stimulating antigen-presenting cells that present autoantigens and enhancing the expression of co-stimulatory molecules and cytokines, thus leading to the loss of self-tolerance and the production of autoantibodies. Additionally, the break of gut barrier and the translocation of gut commensals to inner organs can trigger immune dysregulation and inappropriate systemic inflammation. All these microbiota-mediated mechanisms could contribute to lupus immunopathogenesis and promote disease development in susceptible individuals.
Summary
Evidence of the causative role of disturbed gut microbiome in SLE is still limited, and the related molecular mechanisms and pathways are largely elusive. However, the modification of gut microbiota, such as pathobiont vaccine, special diet, restricted consortium transplantation, as well as regulatory metabolites supplementation, might be promising strategies for lupus prophylaxis and treatment.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
Bach JF. The effect of infections on susceptibility to autoimmune and allergic diseases. N Engl J Med. 2002;347(12):911–20. https://doi.org/10.1056/NEJMra020100.
Gremese E, Alivernini S, Ferraccioli ES, Ferraccioli G. Checkpoint inhibitors (CPI) and autoimmune chronic inflammatory diseases (ACIDs): tolerance and loss of tolerance in the occurrence of immuno-rheumatologic manifestations. Clin Immunol. 2020;214:108395. https://doi.org/10.1016/j.clim.2020.108395.
Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–23. https://doi.org/10.1038/nri2515.
Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. https://doi.org/10.1152/physrev.00045.2009.
Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–73. https://doi.org/10.1126/science.1223490.
Jiao Y, Wu L, Huntington ND, Zhang X. Crosstalk between gut microbiota and innate immunity and its implication in autoimmune diseases. Front Immunol. 2020;11:282. https://doi.org/10.3389/fimmu.2020.00282.
Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell. 2014;157(1):121–41. https://doi.org/10.1016/j.cell.2014.03.011.
Tlaskalová-Hogenová H, Stěpánková R, Kozáková H, Hudcovic T, Vannucci L, Tučková L, et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: contribution of germ-free and gnotobiotic animal models of human diseases. Cell Mol Immunol. 2011;8(2):110–20. https://doi.org/10.1038/cmi.2010.67.
van der Meulen TA, Harmsen HJM, Vila AV, Kurilshikov A, Liefers SC, Zhernakova A, et al. Shared gut, but distinct oral microbiota composition in primary Sjögren’s syndrome and systemic lupus erythematosus. J Autoimmun. 2019;97:77–87. https://doi.org/10.1016/j.jaut.2018.10.009.
Ricciuto A, Sherman PM, Laxer RM. Gut microbiota in chronic inflammatory disorders: a focus on pediatric inflammatory bowel diseases and juvenile idiopathic arthritis. Clin Immunol. 2020;215:108415. https://doi.org/10.1016/j.clim.2020.108415.
Zhang X, Chen BD, Zhao LD, Li H. The gut microbiota: emerging evidence in autoimmune diseases. Trends Mol Med. 2020;26(9):862–73. https://doi.org/10.1016/j.molmed.2020.04.001.
Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. https://doi.org/10.1038/nm.3914.
Zhou C, Zhao H, Xiao XY, Chen BD, Guo RJ, Wang Q, et al. Metagenomic profiling of the pro-inflammatory gut microbiota in ankylosing spondylitis. J Autoimmun. 2020;107:102360. https://doi.org/10.1016/j.jaut.2019.102360.
•• Chen BD, Jia XM, Xu JY, Zhao LD, Ji JY, Wu BX, et al. The gut microbiota of non-treated patients with SLE defines an autoimmunogenic and proinflammatory profile. Arthritis Rheumatol. 2020. https://doi.org/10.1002/art.41511. This is the most recent study digging the altered gut microbiota profile in non-treated SLE patients.
Chen B, Sun L, Zhang X. Integration of microbiome and epigenome to decipher the pathogenesis of autoimmune diseases. J Autoimmun. 2017;83:31–42. https://doi.org/10.1016/j.jaut.2017.03.009.
McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL, Foster KR. Host selection of microbiota via differential adhesion. Cell Host Microbe. 2016;19(4):550–9. https://doi.org/10.1016/j.chom.2016.02.021.
Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science. 2018;360(6390):795–800. https://doi.org/10.1126/science.aaq0926.
Peterson DA, McNulty NP, Guruge JL, Gordon JI. IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell Host Microbe. 2007;2(5):328–39. https://doi.org/10.1016/j.chom.2007.09.013.
Yang C, Mogno I, Contijoch EJ, Borgerding JN, Aggarwala V, Li Z, et al. Fecal IgA levels are determined by strain-level differences in Bacteroides ovatus and are modifiable by gut microbiota manipulation. Cell Host Microbe. 2020;27(3):467–75.e6. https://doi.org/10.1016/j.chom.2020.01.016.
Wesemann DR, Portuguese AJ, Meyers RM, Gallagher MP, Cluff-Jones K, Magee JM, et al. Microbial colonization influences early B-lineage development in the gut lamina propria. Nature. 2013;501(7465):112–5. https://doi.org/10.1038/nature12496.
Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500(7461):232–6. https://doi.org/10.1038/nature12331.
Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331(6015):337–41. https://doi.org/10.1126/science.1198469.
López P, González-Rodríguez I, Gueimonde M, Margolles A, Suárez A. Immune response to Bifidobacterium bifidum strains support Treg/Th17 plasticity. PLoS One. 2011;6(9):e24776. https://doi.org/10.1371/journal.pone.0024776.
Geuking MB, Cahenzli J, Lawson MA, Ng DC, Slack E, Hapfelmeier S, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34(5):794–806. https://doi.org/10.1016/j.immuni.2011.03.021.
Geuking MB, McCoy KD, Macpherson AJ. The continuum of intestinal CD4+ T cell adaptations in host-microbial mutualism. Gut Microbes. 2011;2(6):353–7. https://doi.org/10.4161/gmic.18604.
Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, et al. Th17 cell induction by adhesion of microbes to intestinal epithelial cells. Cell. 2015;163(2):367–80. https://doi.org/10.1016/j.cell.2015.08.058.
Campisi L, Barbet G, Ding Y, Esplugues E, Flavell RA, Blander JM. Apoptosis in response to microbial infection induces autoreactive TH17 cells. Nat Immunol. 2016;17(9):1084–92. https://doi.org/10.1038/ni.3512.
Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28(10):1221–7. https://doi.org/10.1136/gut.28.10.1221.
Ratajczak W, Ryl A, Mizerski A, Walczakiewicz K, Sipak O, Laszczynska M. Immunomodulatory potential of gut microbiome-derived short-chain fatty acids (SCFAs). Acta Biochim Pol. 2019;66(1):1–12. https://doi.org/10.18388/abp.2018_2648.
•• Choi SC, Brown J, Gong M, Ge Y, Zadeh M, Li W, et al. Gut microbiota dysbiosis and altered tryptophan catabolism contribute to autoimmunity in lupus-susceptible mice. Sci Transl Med. 2020;12(551). https://doi.org/10.1126/scitranslmed.aax2220. This study reported the interplay of typtophan metabolism, gut microbiota, and lupus immunopathogenesis, and suggested the potential of diet intervention in lupus control.
•• Ma Y, Xu X, Li M, Cai J, Wei Q, Niu H. Gut microbiota promote the inflammatory response in the pathogenesis of systemic lupus erythematosus. Mol Med. 2019;25(1):35. https://doi.org/10.1186/s10020-019-0102-5. This study indicated a fecal microbiome transplantation could transfer lupus phenotype.
Johnson MB, Gaudreau C-M, Gudi R, Brown R, Gilkeson G, Vasu C. Gut microbiota differently contributes to intestinal immune phenotype and systemic autoimmune progression in female and male lupus-prone mice. J Autoimmun. 108:102420. https://doi.org/10.1016/j.jaut.2020.102420.
Johnson BM, Gaudreau MC, Al-Gadban MM, Gudi R, Vasu C. Impact of dietary deviation on disease progression and gut microbiome composition in lupus-prone SNF1 mice. Clin Exp Immunol. 2015;181(2):323–37. https://doi.org/10.1111/cei.12609.
Zhang H, Liao X, Sparks JB, Luo XM. Dynamics of gut microbiota in autoimmune lupus. Appl Environ Microbiol. 2014;80(24):7551–60. https://doi.org/10.1128/aem.02676-14.
Mu Q, Tavella VJ, Kirby JL, Cecere TE, Chung M, Lee J, et al. Antibiotics ameliorate lupus-like symptoms in mice. Sci Rep. 2017;7(1):13675. https://doi.org/10.1038/s41598-017-14223-0.
Mu Q, Zhang H, Liao X, Lin K, Liu H, Edwards MR, et al. Control of lupus nephritis by changes of gut microbiota. Microbiome. 2017;5(1):73. https://doi.org/10.1186/s40168-017-0300-8.
Luo XM, Edwards MR, Mu Q, Yu Y, Vieson MD, Reilly CM, et al. Gut microbiota in human systemic lupus erythematosus and a mouse model of lupus. Appl Environ Microbiol. 2018;84(4). https://doi.org/10.1128/AEM.02288-17.
•• Zegarra-Ruiz DF, El Beidaq A, Iniguez AJ, Lubrano Di Ricco M, Manfredo Vieira S, Ruff WE, et al. A diet-sensitive commensal Lactobacillus strain mediates TLR7-dependent systemic autoimmunity. Cell Host Microbe. 2019;25(1):113–27 e6. https://doi.org/10.1016/j.chom.2018.11.009. This study identified the pathogenic role of Lactobacillus reuteri in lupus via translocation and type 1 interferon activation.
Manirarora JN, Kosiewicz MM, Alard P. Feeding lactobacilli impacts lupus progression in (NZBxNZW)F1 lupus-prone mice by enhancing immunoregulation. Autoimmunity. 2020;53(6):323–32. https://doi.org/10.1080/08916934.2020.1777282.
Toral M, Robles-Vera I, Romero M, de la Visitación N, Sánchez M, O’Valle F, et al. Lactobacillus fermentum CECT5716: a novel alternative for the prevention of vascular disorders in a mouse model of systemic lupus erythematosus. FASEB J. 2019;33(9):10005–18. https://doi.org/10.1096/fj.201900545RR.
Li Y, Wang HF, Li X, Li HX, Zhang Q, Zhou HW, et al. Disordered intestinal microbes are associated with the activity of systemic lupus erythematosus. Clin Sci. 2019;133(7):821–38. https://doi.org/10.1042/cs20180841.
Azzouz D, Omarbekova A, Heguy A, Schwudke D, Gisch N, Rovin BH, et al. Lupus nephritis is linked to disease-activity associated expansions and immunity to a gut commensal. Ann Rheum Dis. 2019;78(7):947–56. https://doi.org/10.1136/annrheumdis-2018-214856.
Guo M, Wang H, Xu S, Zhuang Y, An J, Su C, et al. Alteration in gut microbiota is associated with dysregulation of cytokines and glucocorticoid therapy in systemic lupus erythematosus. Gut Microbes. 2020;11(6):1758–73. https://doi.org/10.1080/19490976.2020.1768644.
Hevia A, Milani C, Lopez P, Cuervo A, Arboleya S, Duranti S, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. mBio. 2014;5(5):e01548–14. https://doi.org/10.1128/mBio.01548-14.
He Z, Shao T, Li H, Xie Z, Wen C. Alterations of the gut microbiome in Chinese patients with systemic lupus erythematosus. Gut Pathog. 2016;8:64. https://doi.org/10.1186/s13099-016-0146-9.
•• Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science. 2018;359(6380):1156–61. https://doi.org/10.1126/science.aar7201. This study is a milestone in deciphering the autoimmunogenic and proinflammatory role of a gut pathobiont in SLE.
Neuman H, Koren O. The gut microbiota: a possible factor influencing systemic lupus erythematosus. Curr Opin Rheumatol. 2017;29(4):374–7. https://doi.org/10.1097/bor.0000000000000395.
Mu Q, Zhang H, Luo XM. SLE: another autoimmune disorder influenced by microbes and diet? Front Immunol. 2015;6:608. https://doi.org/10.3389/fimmu.2015.00608.
Thim-Uam A, Surawut S, Issara-Amphorn J, Jaroonwitchawan T, Hiengrach P, Chatthanathon P, et al. Leaky-gut enhanced lupus progression in the Fc gamma receptor-IIb deficient and pristane-induced mouse models of lupus. Sci Rep. 2020;10(1):777. https://doi.org/10.1038/s41598-019-57275-0.
Ogunrinde E, Zhou Z, Luo Z, Alekseyenko A, Li QZ, Macedo D, et al. A link between plasma microbial translocation, microbiome, and autoantibody development in first-degree relatives of systemic lupus erythematosus patients. Arthritis Rheumatol. 2019;71(11):1858–68. https://doi.org/10.1002/art.40935.
Deshmukh US, Lewis JE, Gaskin F, Dhakephalkar PK, Kannapell CC, Waters ST, et al. Ro60 peptides induce antibodies to similar epitopes shared among lupus-related autoantigens. J Immunol. 2000;164(12):6655–61. https://doi.org/10.4049/jimmunol.164.12.6655.
Greiling TM, Dehner C, Chen X, Hughes K, Iñiguez AJ, Boccitto M, et al. Commensal orthologs of the human autoantigen Ro60 as triggers of autoimmunity in lupus. Sci Transl Med. 2018;10(434). https://doi.org/10.1126/scitranslmed.aan2306.
Clancy RM, Marion MC, Ainsworth HC, Blaser MJ, Chang M, Howard TD, et al. Salivary dysbiosis and the clinical spectrum in anti-Ro positive mothers of children with neonatal lupus. J Autoimmun. 2020;107:102354. https://doi.org/10.1016/j.jaut.2019.102354.
Zhao Z, Ren J, Dai C, Kannapell CC, Wang H, Gaskin F, et al. Nature of T cell epitopes in lupus antigens and HLA-DR determines autoantibody initiation and diversification. Ann Rheum Dis. 2019;78(3):380–90. https://doi.org/10.1136/annrheumdis-2018-214125.
Pericleous C, D’Souza A, McDonnell T, Ripoll VM, Leach O, Isenberg D, et al. Antiphospholipid antibody levels in early systemic lupus erythematosus: are they associated with subsequent mortality and vascular events? Rheumatology. 2020;59(1):146–52. https://doi.org/10.1093/rheumatology/kez239.
Ruff WE, Dehner C, Kim WJ, Pagovich O, Aguiar CL, Yu AT, et al. Pathogenic autoreactive T and B cells cross-react with mimotopes expressed by a common human gut commensal to trigger autoimmunity. Cell Host Microbe. 2019;26(1):100–13 e8. https://doi.org/10.1016/j.chom.2019.05.003.
López P, de Paz B, Rodríguez-Carrio J, Hevia A, Sánchez B, Margolles A, et al. Th17 responses and natural IgM antibodies are related to gut microbiota composition in systemic lupus erythematosus patients. Sci Rep. 2016;6:24072. https://doi.org/10.1038/srep24072.
Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504(7480):451–5. https://doi.org/10.1038/nature12726.
Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341(6145):569–73. https://doi.org/10.1126/science.1241165.
Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T, et al. The effect of short-chain fatty acids on human monocyte-derived dendritic cells. Sci Rep. 2015;5:16148. https://doi.org/10.1038/srep16148.
•• Sanchez HN, Moroney JB, Gan H, Shen T, Im JL, Li T, et al. B cell-intrinsic epigenetic modulation of antibody responses by dietary fiber-derived short-chain fatty acids. Nat Commun. 2020;11(1):60. https://doi.org/10.1038/s41467-019-13603-6. This study revealed the impact of SCFA on B cell intrinsic function and the involved pathways and molecules.
Crow MK. Type I interferon in the pathogenesis of lupus. J Immunol. 2014;192(12):5459–68. https://doi.org/10.4049/jimmunol.1002795.
Mu Q, Cabana-Puig X, Mao J, Swartwout B, Abdelhamid L, Cecere TE, et al. Pregnancy and lactation interfere with the response of autoimmunity to modulation of gut microbiota. Microbiome. 2019;7(1):105. https://doi.org/10.1186/s40168-019-0720-8.
Lippens C, Duraes FV, Dubrot J, Brighouse D, Lacroix M, Irla M, et al. IDO-orchestrated crosstalk between pDCs and Tregs inhibits autoimmunity. J Autoimmun. 2016;75:39–49. https://doi.org/10.1016/j.jaut.2016.07.004.
Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139(3):485–98. https://doi.org/10.1016/j.cell.2009.09.033.
Gaboriau-Routhiau V, Rakotobe S, Lécuyer E, Mulder I, Lan A, Bridonneau C, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31(4):677–89. https://doi.org/10.1016/j.immuni.2009.08.020.
Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, et al. Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity. 2010;32(6):815–27. https://doi.org/10.1016/j.immuni.2010.06.001.
Van Praet JT, Donovan E, Vanassche I, Drennan MB, Windels F, Dendooven A, et al. Commensal microbiota influence systemic autoimmune responses. EMBO J. 2015;34(4):466–74. https://doi.org/10.15252/embj.201489966.
Funding
This study was supported by grants from the National Natural Science Foundation of China (81788101, 81630044, 81771763, 82071840), Chinese Academy of Medical Science Innovation Fund for Medical Sciences (CIFMS2017-12M-1-008, 2016-12M-1-003, 2017-I2M-3-011, 2016-12M-1-008, 2020-12M-C&T-B-013), Beijing Capital Health Development Fund (2020-2-4019), and Grant from Medical Epigenetics Research Center, Chinese Academy of Medical Sciences (2017PT31035).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topical Collection on Systemic Lupus Erythematosus
Rights and permissions
About this article
Cite this article
Li, R., Meng, X., Chen, B. et al. Gut Microbiota in Lupus: a Butterfly Effect?. Curr Rheumatol Rep 23, 27 (2021). https://doi.org/10.1007/s11926-021-00986-z
Accepted:
Published:
DOI: https://doi.org/10.1007/s11926-021-00986-z