Abstract
Purpose of Review
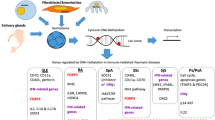
DNA methylation has emerged as an important contributing factor in the pathogenesis of systemic lupus erythematosus (SLE). Here, we describe the DNA methylation patterns identified in SLE and how these epigenetic changes can influence disease susceptibility, clinical heterogeneity, and disease flares.
Recent Findings
Several genome-wide DNA methylation studies have been recently completed in SLE. Important observations include robust demethylation of interferon-regulated genes, which is consistent across all cell types studied to date, and is independent of disease activity. This interferon epigenetic signature was shown to precede interferon transcription signature in SLE, suggesting it might be an early event in the disease process. Recent studies also revealed DNA methylation changes specific for renal and skin involvement in SLE, providing a proof of principle for a value of DNA methylation studies in exploring mechanisms of specific disease manifestations, and potentially as prognostic biomarkers. Inherited ethnicity-specific DNA methylation patterns have also been shown to possibly contribute to differences in SLE susceptibility between populations. Finally, a recent study revealed that DNA methylation levels at IFI44L can accurately distinguish SLE patients from healthy controls, and from patients with other autoimmune diseases, promising to be the first epigenetic diagnostic marker for SLE.
Summary
Genome-wide DNA methylation studies in SLE have provided novel insights into disease pathogenesis, clinical heterogeneity, and disease flares. Further studies promise to reveal novel diagnostic, prognostic, and therapeutic targets for SLE.




Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance, •• Of major importance
Ballestar E. Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol. 2011;7:263–71.
Gupta B, Hawkins RD. Epigenomics of autoimmune diseases. Immunol Cell Biol. 2015;93:271–6.
Feinberg AP, Koldobskiy MA, Gondor A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016;17:284–99.
Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27.
Jeffries MA, Sawalha AH. Autoimmune disease in the epigenetic era: how has epigenetics changed our understanding of disease and how can we expect the field to evolve? Expert Rev Clin Immunol. 2015;11:45–58.
•• Farh KK, Marson A, Zhu J, Kleinewietfeld M, Housley WJ, Beik S, Shoresh N, Whitton H, Ryan RJ, Shishkin AA, et al. Genetic and epigenetic fine mapping of causal autoimmune disease variants. Nature. 2015;518:337–43. The aim of the study is to identify the causality of the genome-wide association variants in autoimmune diseases using epigenetic marks. The study found that ~90% of causal variants are non-coding, with ~60% mapping to immune-cell enhancers and only 10–20% directly alter recognizable transcription factor binding motifs
Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17:630–41.
• Teruel M, Alarcon-Riquelme ME. The genetic basis of systemic lupus erythematosus: what are the risk factors and what have we learned. J Autoimmun. 2016;74:161–75. Recent review about the genetic risk factors to SLE susceptibility identified across populations
Deng Y, Tsao BP. Genetic susceptibility to systemic lupus erythematosus in the genomic era. Nat Rev Rheumatol. 2010;6:683–92.
Deng Y, Tsao BP. Advances in lupus genetics and epigenetics. Curr Opin Rheumatol. 2014;26:482–92.
Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22.
Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 1990;33:1665–73.
Quddus J, Johnson KJ, Gavalchin J, Amento EP, Chrisp CE, Yung RL, Richardson BC. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53.
Yung RL, Quddus J, Chrisp CE, Johnson KJ, Richardson BC. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J Immunol. 1995;154:3025–35.
Richardson B, Sawalha AH, Ray D, Yung R. Murine models of lupus induced by hypomethylated T cells (DNA hypomethylation and lupus...). Methods Mol Biol. 2012;900:169–80.
Lu Q, Wu A, Richardson BC. Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J Immunol. 2005;174:6212–9.
Lu Q, Kaplan M, Ray D, Ray D, Zacharek S, Gutsch D, Richardson B. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 2002;46:1282–91.
Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J Immunol. 2007;179:6352–8.
Sawalha AH, Jeffries M, Webb R, Lu Q, Gorelik G, Ray D, Osban J, Knowlton N, Johnson K, Richardson B. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 2008;9:368–78.
Gorelik G, Richardson B. Key role of ERK pathway signaling in lupus. Autoimmunity. 2010;43:17–22.
Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O'Hanlon TP, Rider LG, et al. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20:170–9.
Jeffries MA, Dozmorov M, Tang Y, Merrill JT, Wren JD, Sawalha AH. Genome-wide DNA methylation patterns in CD4+ T cells from patients with systemic lupus erythematosus. Epigenetics. 2011;6:593–601.
Li Y, Liu Y, Strickland FM, Richardson B. Age-dependent decreases in DNA methyltransferase levels and low transmethylation micronutrient levels synergize to promote overexpression of genes implicated in autoimmunity and acute coronary syndromes. Exp Gerontol. 2010;45:312–22.
Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, Merrill JT, McCune WJ, Sawalha AH. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. J Autoimmun. 2013;43:78–84.
Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678.
Coit P, Yalavarthi S, Ognenovski M, Zhao W, Hasni S, Wren JD, Kaplan MJ, Sawalha AH. Epigenome profiling reveals significant DNA demethylation of interferon signature genes in lupus neutrophils. J Autoimmun. 2015;58:59–66.
Knight JS, Kaplan MJ. Lupus neutrophils: ‘NET’ gain in understanding lupus pathogenesis. Curr Opin Rheumatol. 2012;24:441–50.
Sukapan P, Promnarate P, Avihingsanon Y, Mutirangura A, Hirankarn N. Types of DNA methylation status of the interspersed repetitive sequences for LINE-1, Alu, HERV-E and HERV-K in the neutrophils from systemic lupus erythematosus patients and healthy controls. J Hum Genet. 2014;59:178–88.
Mavragani CP, Sagalovskiy I, Guo Q, Nezos A, Kapsogeorgou EK, Lu P, Liang Zhou J, Kirou KA, Seshan SV, Moutsopoulos HM, et al. Expression of long interspersed nuclear element 1 retroelements and induction of type I interferon in patients with systemic autoimmune disease. Arthritis & rheumatology. 2016;68:2686–96.
•• Coit P, Dozmorov MG, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Wren JD, Sawalha AH. Epigenetic reprogramming in naive CD4+ T cells favoring T cell activation and non-Th1 effector T cell immune response as an early event in lupus flares. Arthritis & rheumatology. 2016;68:2200–9. The first genome-wide DNA methylation analysis in naïve CD4 + T cells from SLE patients with different degrees of disease activity in order to investigate the earliest T-cell epigenetic change associated with SLE disease flares
Zhao E, Maj T, Kryczek I, Li W, Wu K, Zhao L, Wei S, Crespo J, Wan S, Vatan L, et al. Cancer mediates effector T cell dysfunction by targeting microRNAs and EZH2 via glycolysis restriction. Nat Immunol. 2016;17:95–103.
Yin Y, Choi SC, Xu Z, Perry DJ, Seay H, Croker BP, Sobel ES, Brusko TM, Morel L. Normalization of CD4+ T cell metabolism reverses lupus. Sci Transl Med. 2015;7:274ra218.
Gonzalez LA, Toloza SM, Alarcon GS. Impact of race and ethnicity in the course and outcome of systemic lupus erythematosus. Rheum Dis Clin N Am. 2014;40:433–54. vii-viii
Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+ T cells. J Autoimmun. 2015;61:29–35.
Mok A, Solomon O, Nayak RR, Coit P, Quach HL, Nititham J, Sawalha AH, Barcellos LF, Criswell LA, Chung SA. Genome-wide profiling identifies associations between lupus nephritis and differential methylation of genes regulating tissue hypoxia and type 1 interferon responses. Lupus science & medicine. 2016;3:e000183.
International Consortium for Systemic Lupus Erythematosus, G, Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–10.
Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, Zhang Q, Zhang P, Yu X, Xia Y, et al. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. J Autoimmun. 2014;54:127–36.
Fu Q, Zhao J, Qian X, Wong JL, Kaufman KM, Yu CY, Hwee Siew H, Tan Tock Seng Hospital Lupus Study, G, Mok MY, Harley JB, et al. Association of a functional IRF7 variant with systemic lupus erythematosus. Arthritis Rheum. 2011;63:749–54.
Kawasaki A, Furukawa H, Kondo Y, Ito S, Hayashi T, Kusaoi M, Matsumoto I, Tohma S, Takasaki Y, Hashimoto H, et al. Association of PHRF1-IRF7 region polymorphism with clinical manifestations of systemic lupus erythematosus in a Japanese population. Lupus. 2012;21:890–5.
Renauer P, Coit P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. DNA methylation patterns in naive CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus science & medicine. 2015;2:e000101.
Chung SA, Nititham J, Elboudwarej E, Quach HL, Taylor KE, Barcellos LF, Criswell LA. Genome-wide assessment of differential DNA methylation associated with autoantibody production in systemic lupus erythematosus. PLoS One. 2015;10:e0129813.
Yu C, Gershwin ME, Chang C. Diagnostic criteria for systemic lupus erythematosus: a critical review. J Autoimmun. 2014;48-49:10–3.
Hughes T, Adler A, Merrill JT, Kelly JA, Kaufman KM, Williams A, Langefeld CD, Gilkeson GS, Sanchez E, Martin J, et al. Analysis of autosomal genes reveals gene-sex interactions and higher total genetic risk in men with systemic lupus erythematosus. Ann Rheum Dis. 2012;71:694–9.
Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, Chen L, Cunninghame Graham DS, Bentham J, Roberts AL, et al. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet. 2016;48:940–6.
Taylor KE, Chung SA, Graham RR, Ortmann WA, Lee AT, Langefeld CD, Jacob CO, Kamboh MI, Alarcon-Riquelme ME, Tsao BP, et al. Risk alleles for systemic lupus erythematosus in a large case-control collection and associations with clinical subphenotypes. PLoS Genet. 2011;7:e1001311.
Heyn H, Esteller M. DNA methylation profiling in the clinic: applications and challenges. Nat Rev Genet. 2012;13:679–92.
•• Zhao M, Zhou Y, Zhu B, Wan M, Jiang T, Tan Q, Liu Y, Jiang J, Luo S, Tan Y, et al. IFI44L promoter methylation as a blood biomarker for systemic lupus erythematosus. Ann Rheum Dis. 2016;75:1998–2006. First genome-wide DNA methylation study in whole blood to investigate whether specific gene methylation changes could meet sensitivity and specificity criteria for a robust diagnostic biomarker in SLE. The methylation level of IFI44L promoter was found to distinguish patients with SLE from healthy persons and other autoimmune diseases, and was a highly sensitive and specific diagnostic marker for SLE, suggesting this epigenetic mark as potential diagnostic biomarker of SLE
Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, Alarcon-Riquelme ME, James JA, Vyse TJ, Gilkeson GS, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS One. 2008;3:e1727.
Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, Frank MB, Merrill J, Kimberly RP, Edberg JC, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60:1076–84.
Kaufman KM, Zhao J, Kelly JA, Hughes T, Adler A, Sanchez E, Ojwang JO, Langefeld CD, Ziegler JT, Williams AH, et al. Fine mapping of Xq28: both MECP2 and IRAK1 contribute to risk for systemic lupus erythematosus in multiple ancestral groups. Ann Rheum Dis. 2013;72:437–44.
Koelsch KA, Webb R, Jeffries M, Dozmorov MG, Frank MB, Guthridge JM, James JA, Wren JD, Sawalha AH. Functional characterization of the MECP2/IRAK1 lupus risk haplotype in human T cells and a human MECP2 transgenic mouse. J Autoimmun. 2013;41:168–74.
Zhang FF, Cardarelli R, Carroll J, Fulda KG, Kaur M, Gonzalez K, Vishwanatha JK, Santella RM, Morabia A. Significant differences in global genomic DNA methylation by gender and race/ethnicity in peripheral blood. Epigenetics. 2011;6:623–9.
Heyn H, Moran S, Hernando-Herraez I, Sayols S, Gomez A, Sandoval J, Monk D, Hata K, Marques-Bonet T, Wang L, et al. DNA methylation contributes to natural human variation. Genome Res. 2013;23:1363–72.
Coit P, Ognenovski M, Gensterblum E, Maksimowicz-McKinnon K, Wren JD, Sawalha AH. Ethnicity-specific epigenetic variation in naive CD4+ T cells and the susceptibility to autoimmunity. Epigenetics Chromatin. 2015;8:49.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award number R01AI097134 and award number U19AI110502. M.T. is supported by the Instituto de Salud Carlos III from Spain through the Sara Borrell subprogram (CD13/00316).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
A.H.S. is listed as inventor on a patent application to use IFI44L methylation as a diagnostic test for lupus.
M.T. declares no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Systemic Lupus Erythematosus
Rights and permissions
About this article
Cite this article
Teruel, M., Sawalha, A.H. Epigenetic Variability in Systemic Lupus Erythematosus: What We Learned from Genome-Wide DNA Methylation Studies. Curr Rheumatol Rep 19, 32 (2017). https://doi.org/10.1007/s11926-017-0657-5
Published:
DOI: https://doi.org/10.1007/s11926-017-0657-5