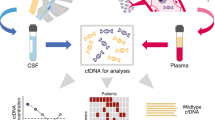
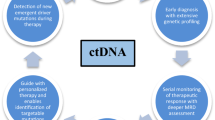
Abstract
Purpose of Review
Recent advances have been made in circulating tumor DNA (ctDNA), the method to minimally invasive detect lymphoma sensitively with tumor-derived DNA in the blood of patients with lymphomas. This article discusses these various methods of ctDNA detection and the clinical context in which they have been applied to for a variety of lymphoma subtypes.
Recent Findings
ctDNA has been applied to a variety of subtypes of lymphoma and has been used in the context of genotyping somatic mutations and classification of disease, monitoring of response during treatment, detecting minimal residual disease even with radiographic remission, and predicting relapse and long-term survival outcomes. There are a variety of techniques used to measure ctDNA including digital polymerase chain reaction and next-generation sequencing techniques including high-throughput variable-diversity-joining rearrangement sequencing, high-throughput sequencing of somatic mutations, and Cancer Personalized Profiling by deep sequencing. While the greatest data has been generated in diffuse large B cell lymphoma, there have been studies utilizing application of ctDNA in follicular lymphoma, mantle cell lymphoma, Hodgkin’s lymphoma, peripheral T cell lymphoma, and primary CNS lymphoma among others.
Summary
ctDNA is an emerging biomarker in lymphoma that can minimally invasively provide further genotypic information, diagnostic clarification, and treatment prognostication by detection of minimal residual disease even without radiographic evidence of disease. Future studies are needed to standardize the use of ctDNA and translate its use clinically for the management of lymphoma patients.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Roschewski M, Rossi D, Kurtz DM, et al. Circulating tumor DNA in lymphoma: principles and future directions. Blood Cancer Discov. 2022;3:5–15. These references are key papers that review the general principles of ctDNA, methods of ctDNA measurement, and key clinical applications of ctDNA with respect to prognosis and early detection in lymphoma.
Hindson CM, Chevillet JR, Briggs HA, et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat Methods. 2013;10:1003–5.
Huet S, Salles G. Potential of circulating tumor DNA for the management of patients with lymphoma. JCO Oncol Pract. 2020;16:561–568. These references are key papers that review the general principles of ctDNA, methods of ctDNA measurement, and key clinical applications of ctDNA with respect to prognosis and early detection in lymphoma.
Camus V, Jardin F. Cell-free DNA and the monitoring of lymphoma treatment. Pharmacogenomics. 2019;20:1271–82.
Jung D, Jain P, Yao Y, et al. Advances in the assessment of minimal residual disease in mantle cell lymphoma. J Hematol Oncol. 2020;13:127. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Hussaini MO, Srivastava J, Lee LW, et al. Moffitt cancer center 2-year single-institution experience with next-generation sequencing minimal residual disease detection: clinical utility, application, and correlation with outcomes in plasma cell and lymphoid malignancies. Blood. 2019;134(Supplement_1):4654-.
Dubois S, Viailly PJ, Mareschal S, et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: a LYSA study. Clin Cancer Res. 2016;22:2919–28. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Bohers E, Viailly PJ, Becker S, et al. Non-invasive monitoring of diffuse large B-cell lymphoma by cell-free DNA high-throughput targeted sequencing: analysis of a prospective cohort. Blood Cancer J. 2018;8:74. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Scherer F, Kurtz DM, Newman AM, et al. Distinct biological subtypes and patterns of genome evolution in lymphoma revealed by circulating tumor DNA. Sci Transl Med. 2016;8:364ra155.
Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014;32:579–86.
Rossi D, Spina V, Bruscaggin A, et al. Liquid biopsy in lymphoma. Haematologica. 2019;104:648-652. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Bratman SV, Newman AM, Alizadeh AA, et al. Potential clinical utility of ultrasensitive circulating tumor DNA detection with CAPP-Seq. Expert Rev Mol Diagn. 2015;15:715–9.
Roschewski M, Dunleavy K, Pittaluga S, et al. Circulating tumour DNA and CT monitoring in patients with untreated diffuse large B-cell lymphoma: a correlative biomarker study. Lancet Oncol. 2015;16:541–9. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Kurtz DM, Green MR, Bratman SV, et al. Noninvasive monitoring of diffuse large B-cell lymphoma by immunoglobulin high-throughput sequencing. Blood. 2015;125:3679–87. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Alig S, Macaulay CW, Kurtz DM, et al: Short diagnosis-to-treatment interval is associated with higher circulating tumor DNA levels in diffuse large B-cell lymphoma. J Clin Oncol. 2021;39:2605–2616. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Kurtz DM, Scherer F, Jin MC, et al. Circulating tumor DNA measurements as early outcome predictors in diffuse large B-cell lymphoma. J Clin Oncol. 2018;36:2845–2853. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Herrera AF, Tracy S, Croft B, et al. Risk profiling of patients with relapsed/refractory diffuse large B-cell lymphoma by measuring circulating tumor DNA. Blood Adv. 2022;6(6):1651–60.
Frank MJ, Hossain NM, Bukhari A, et al. Monitoring of circulating tumor DNA improves early relapse detection after axicabtagene ciloleucel infusion in large B-cell lymphoma: results of a prospective multi-institutional trial. J Clin Oncol. 2021;39:3034–3043. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Cirillo M, Craig AFM, Borchmann S, et al. Liquid biopsy in lymphoma: molecular methods and clinical applications. Cancer Treat Rev. 2020;91:102106.
Rossi D, Diop F, Spaccarotella E, et al. Diffuse large B-cell lymphoma genotyping on the liquid biopsy. Blood. 2017;129:1947–1957. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Camus V, Bohers E, Dubois S, et al. Circulating tumor DNA: an important tool in precision medicine for lymphoma. Expert Rev Precis Med Drug Dev. 2018;3:11–21. These references are key papers that review the general principles of ctDNA, methods of ctDNA measurement, and key clinical applications of ctDNA with respect to prognosis and early detection in lymphoma.
Kumar A, Westin J, Schuster SJ, et al. Interim analysis from a prospective multicenter study of next-generation sequencing minimal residual disease assessment and ct monitoring for surveillance after frontline treatment in diffuse large b-cell lymphoma. Blood. 2020;136(Supplement 1):46–7.
Kurtz DM, Soo J, Co Ting Keh L, et al. Enhanced detection of minimal residual disease by targeted sequencing of phased variants in circulating tumor DNA. Nat Biotechnol. 2021;39:1537–1547.
Distler A, Lakhotia R, Phelan JD, et al. A prospective study of clonal evolution in follicular lymphoma: circulating tumor DNA correlates with overall tumor burden and fluctuates over time without therapy. Blood. 2021;138:1328–1328. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Sarkozy C, Huet S, Carlton VE, et al. The prognostic value of clonal heterogeneity and quantitative assessment of plasma circulating clonal IG-VDJ sequences at diagnosis in patients with follicular lymphoma. Oncotarget. 2017;8:8765–74.
Delfau-Larue MH, van der Gucht A, Dupuis J, et al. Total metabolic tumor volume, circulating tumor cells, cell-free DNA: distinct prognostic value in follicular lymphoma. Blood Adv. 2018;2:807–16.
Galimberti S, Luminari S, Ciabatti E, et al. Minimal residual disease after conventional treatment significantly impacts on progression-free survival of patients with follicular lymphoma: the FIL FOLL05 trial. Clin Cancer Res. 2014;20:6398–405. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Sarkozy C, Trneny M, Xerri L, et al. Risk factors and outcomes for patients with follicular lymphoma who had histologic transformation after response to first-line immunochemotherapy in the PRIMA trial. J Clin Oncol. 2016;34:2575–82.
Merryman RW, Edwin N, Redd R, et al. Rituximab/bendamustine and rituximab/cytarabine induction therapy for transplant-eligible mantle cell lymphoma. Blood Adv. 2020;4:858–67.
Smith MR, Jegede O, Martin P, et al. ECOG-ACRIN E1411 randomized phase 2 trial of bendamustine-rituximab (BR)-based induction followed by rituximab (R) ± lenalidomide (L) consolidation for mantle cell lymphoma: effect of adding bortezomib to front-line BR induction on PFS. J Clin Oncol. 2021;39:7503–7503.
Lakhotia R, Melani C, Dunleavy K, Pittaluga S, Saba N, Lindenberg L, Mena E, Bergvall E, Lucas AN, Jacob A, Yusko E, Steinberg SM, Jaffe ES, Wiestner A, Wilson WH, Roschewski M. Circulating tumor DNA predicts therapeutic outcome in mantle cell lymphoma. Blood Adv. 2022;6(8):2667–2680. https://doi.org/10.1182/bloodadvances.2021006397
Kumar A, Bantilan KS, Jacob AP, et al. Noninvasive monitoring of mantle cell lymphoma by immunoglobulin gene next-generation sequencing in a phase 2 study of sequential chemoradioimmunotherapy followed by autologous stem-cell rescue. Clin Lymphoma Myeloma Leuk. 2021;21:230–237 e12.
Camus V, Stamatoullas A, Mareschal S, et al. Detection and prognostic value of recurrent exportin 1 mutations in tumor and cell-free circulating DNA of patients with classical Hodgkin lymphoma. Haematologica. 2016;101:1094–101.
Primerano S, Burnelli R, Carraro E, et al. Kinetics of circulating plasma cell-free DNA in paediatric classical Hodgkin lymphoma. J Cancer. 2016;7:364–6.
Vandenberghe P, Wlodarska I, Tousseyn T, et al. Non-invasive detection of genomic imbalances in Hodgkin/Reed-Sternberg cells in early and advanced stage Hodgkin's lymphoma by sequencing of circulating cell-free DNA: a technical proof-of-principle study. Lancet Haematol. 2015;2:e55–65. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Spina V, Bruscaggin A, Cuccaro A, et al. Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma. Blood. 2018;131:2413–25.
Zhang W, Wang W, Han X, et al. Circulating tumor DNA by high-throughput sequencing of T cell receptor monitored treatment response and predicted treatment failure in T cell lymphomas. Int J Lab Hematol. 2021;43:1041–9.
Miljkovic MD, Melani C, Pittaluga S, et al. Next-generation sequencing-based monitoring of circulating tumor DNA reveals clonotypic heterogeneity in untreated PTCL. Blood Adv. 2021;5:4198–4210. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Fontanilles M, Marguet F, Bohers E, et al. Non-invasive detection of somatic mutations using next-generation sequencing in primary central nervous system lymphoma. Oncotarget. 2017;8:48157–68.
Rimelen V, Ahle G, Pencreach E, et al. Tumor cell-free DNA detection in CSF for primary CNS lymphoma diagnosis. Acta Neuropathol Commun. 2019;7:43.
Watanabe, Jun et al. High detection rate of myd88 mutations in cerebrospinal fluid from patients with cns lymphomas. JCO Precs Oncol. 2019;3:1–13. https://doi.org/10.1200/PO.18.00308
Bobillo S, Crespo M, Escudero L, et al. Cell free circulating tumor DNA in cerebrospinal fluid detects and monitors central nervous system involvement of B-cell lymphomas. Haematologica. 2021;106:513–21.
Mutter JA, et al. Profiling of circulating tumor DNA for noninvasive disease detection, risk stratification, and MRD monitoring in patients with CNS lymphoma. Blood. 2021;138(Supplement 1):6–6. These references are key papers pertaining to the application of ctDNA to specific lymphoma subtypes namely DLBCL, FL, MCL, HL and PTCL.
Johnson P, Federico M, Kirkwood A, et al. Adapted treatment guided by Interim PET-CT scan in advanced Hodgkin’s lymphoma. N Engl J Med. 2016;374:2419–29.
Persky DO, Li H, Stephens DM, et al. Positron emission tomography-directed therapy for patients with limited-stage diffuse large B-cell lymphoma: results of Intergroup National Clinical Trials Network Study S1001. J Clin Oncol. 2020;38:3003–11.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
S.K.: none; J.Z.: Secura Bio, DaichiSankyo, Abbvie: research funding; Kiyoaw Kirin, Secura Bio, Seattle Genetics: honoraria; Secura Bio, Ono, Legend, Kiyowa Kirin, Myeloid Therapeutics Verastem Daichi Sankyo: consultancy.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on T-Cell and Other Lymphoproliferative Malignancies
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kambhampati, S., Zain, J. Circulating Tumor DNA in Lymphoma. Curr Hematol Malig Rep 17, 298–305 (2022). https://doi.org/10.1007/s11899-022-00677-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-022-00677-1