Abstract
Purpose of Review
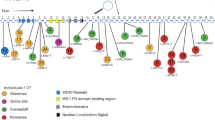
Somatic mutations in DNA methyltransferases and other DNA methylation associated genes have been found in a wide variety of cancers. Germline mutations in these genes have been associated with several rare hereditary disorders. Among the described germline/congenital disorders, neurological dysfunction and/or growth abnormalities appear to be a common phenotype. Here, we outline known germline abnormalities and examine the cancer risks associated with these mutations.
Recent Findings
The increased use and availability of sequencing techniques in the clinical setting has expanded the identification of germline abnormalities involving DNA methylation machinery. This has provided additional cases to study these rare hereditary disorders and their predisposition to cancer.
Summary
Studying these syndromes may offer an opportunity to better understand the contribution of these genes in cancer development.





Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500.
Handy DE, Castro R, Loscalzo J. Epigenetic modifications: basic mechanisms and role in cardiovascular disease. Circulation. 2011;123(19):2145–56.
Song J, Teplova M, Ishibe-Murakami S, Patel DJ. Structure-based mechanistic insights into DNMT1-mediated maintenance DNA methylation. Science. 2012;335(6069):709–12.
Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988;2(9):1136–43.
Panning B, Jaenisch R. RNA and the epigenetic regulation of X chromosome inactivation. Cell. 1998;93(3):305–8.
Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31(2):89–97.
Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304.
Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 2008;27(20):2681–90.
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S, et al. Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol. 2003;13(14):1192–200.
Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, et al. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16(3):304–11.
Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, et al. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet. 1998;19(2):187–91.
Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, et al. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393(6683):386–9.
Song CX, He C. Potential functional roles of DNA demethylation intermediates. Trends Biochem Sci. 2013;38(10):480–4.
Lio CJ, Yue X, Lopez-Moyado IF, Tahiliani M, Aravind L, Rao A. TET methylcytosine oxidases: new insights from a decade of research. J Biosci. 2020;45:21.
He YF, Li BZ, Li Z, Liu P, Wang Y, Tang Q, et al. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science. 2011;333(6047):1303–7.
Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31(1):27–36.
Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–28.
Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–92.
Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–63.
Chedin F, Lieber MR, Hsieh CL. The DNA methyltransferase-like protein DNMT3L stimulates de novo methylation by Dnmt3a. Proc Natl Acad Sci U S A. 2002;99(26):16916–21.
Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–46.
Baylin SB, Jones PA. Epigenetic determinants of cancer. Cold Spring Harb Perspect Biol. 2016;8(9):a019505. https://doi.org/10.1101/cshperspect.a019505.
Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7(1):21–33.
Mikeska T, Craig JM. DNA methylation biomarkers: cancer and beyond. Genes (Basel). 2014;5(3):821–64.
Leygo C, Williams M, Jin HC, Chan MWY, Chu WK, Grusch M, et al. DNA methylation as a noninvasive epigenetic biomarker for the detection of cancer. Dis Markers. 2017;2017:3726595.
Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–56.
Jones PA, Issa JP, Baylin S. Targeting the cancer epigenome for therapy. Nat Rev Genet. 2016;17(10):630–41.
Blecua P, Martinez-Verbo L, Esteller M. The DNA methylation landscape of hematological malignancies: an update. Mol Oncol. 2020;14(8):1616–39.
Hoang NM, Rui L. DNA methyltransferases in hematological malignancies. J Genet Genomics. 2020;47(7):361–72.
Venugopal K, Feng Y, Shabashvili D, Guryanova OA. Alterations to DNMT3A in hematologic malignancies. Can Res. 2021;81(2):254–63.
Norvil AB, Saha D, Dar MS, Gowher H. Effect of disease-associated germline mutations on structure function relationship of DNA methyltransferases. Genes (Basel). 2019;10(5):369. https://doi.org/10.3390/genes10050369.
Klein CJ, Botuyan MV, Wu Y, Ward CJ, Nicholson GA, Hammans S, et al. Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet. 2011;43(6):595–600.
Klein CJ, Bird T, Ertekin-Taner N, Lincoln S, Hjorth R, Wu Y, et al. DNMT1 mutation hot spot causes varied phenotypes of HSAN1 with dementia and hearing loss. Neurology. 2013;80(9):824–8.
Wright A, Dyck PJ. Hereditary sensory neuropathy with sensorineural deafness and early-onset dementia. Neurology. 1995;45(3 Pt 1):560–2.
Hojo K, Imamura T, Takanashi M, Ishii K, Sasaki M, Imura S, et al. Hereditary sensory neuropathy with deafness and dementia: a clinical and neuroimaging study. Eur J Neurol. 1999;6(3):357–61.
Pedroso JL, Povoas Barsottini OG, Lin L, Melberg A, Oliveira AS, Mignot E. A novel de novo exon 21 DNMT1 mutation causes cerebellar ataxia, deafness, and narcolepsy in a Brazilian patient. Sleep. 2013;36(8):1257-9.9A.
Moghadam KK, Pizza F, La Morgia C, Franceschini C, Tonon C, Lodi R, et al. Narcolepsy is a common phenotype in HSAN IE and ADCA-DN. Brain. 2014;137(Pt 6):1643–55.
Winkelmann J, Lin L, Schormair B, Kornum BR, Faraco J, Plazzi G, et al. Mutations in DNMT1 cause autosomal dominant cerebellar ataxia, deafness and narcolepsy. Hum Mol Genet. 2012;21(10):2205–10.
• Tatton-Brown K, Seal S, Ruark E, Harmer J, Ramsay E, Del Vecchio Duarte S, et al. Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat Genet. 2014;46(4):385–8. Initial report of Tatton-Brown-Rahman Syndrome.
Jeffries AR, Maroofian R, Salter CG, Chioza BA, Cross HE, Patton MA, et al. Growth disrupting mutations in epigenetic regulatory molecules are associated with abnormalities of epigenetic aging. Genome Res. 2019;29(7):1057–66.
Shen W, Heeley JM, Carlston CM, Acuna-Hidalgo R, Nillesen WM, Dent KM, et al. The spectrum of DNMT3A variants in Tatton-Brown-Rahman syndrome overlaps with that in hematologic malignancies. Am J Med Genet A. 2017;173(11):3022–8.
Kosaki R, Terashima H, Kubota M, Kosaki K. Acute myeloid leukemia-associated DNMT3A p.Arg882His mutation in a patient with Tatton-Brown-Rahman overgrowth syndrome as a constitutional mutation. Am J Med Genet A. 2017;173(1):250–3.
Hollink I, van den Ouweland AMW, Beverloo HB, Arentsen-Peters S, Zwaan CM, Wagner A. Acute myeloid leukaemia in a case with Tatton-Brown-Rahman syndrome: the peculiar DNMT3A R882 mutation. J Med Genet. 2017;54(12):805–8.
Balci TB, Strong A, Kalish JM, Zackai E, Maris JM, Reilly A, et al. Tatton-Brown-Rahman syndrome: six individuals with novel features. Am J Med Genet A. 2020;182(4):673–80.
Tenorio J, Alarcon P, Arias P, Dapia I, Garcia-Minaur S, Palomares Bralo M, et al. Further delineation of neuropsychiatric findings in Tatton-Brown-Rahman syndrome due to disease-causing variants in DNMT3A: seven new patients. Eur J Hum Genet. 2020;28(4):469–79.
Remacha L, Curras-Freixes M, Torres-Ruiz R, Schiavi F, Torres-Perez R, Calsina B, et al. Gain-of-function mutations in DNMT3A in patients with paraganglioma. Genet Med. 2018;20(12):1644–51.
Heyn P, Logan CV, Fluteau A, Challis RC, Auchynnikava T, Martin CA, et al. Gain-of-function DNMT3A mutations cause microcephalic dwarfism and hypermethylation of Polycomb-regulated regions. Nat Genet. 2019;51(1):96–105.
Hagleitner MM, Lankester A, Maraschio P, Hulten M, Fryns JP, Schuetz C, et al. Clinical spectrum of immunodeficiency, centromeric instability and facial dysmorphism (ICF syndrome). J Med Genet. 2008;45(2):93–9.
Weemaes CM, van Tol MJ, Wang J, van Ostaijen-ten Dam MM, van Eggermond MC, Thijssen PE, et al. Heterogeneous clinical presentation in ICF syndrome: correlation with underlying gene defects. Eur J Hum Genet. 2013;21(11):1219–25.
Xu GL, Bestor TH, Bourc’his D, Hsieh CL, Tommerup N, Bugge M, et al. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402(6758):187–91.
Yehezkel S, Segev Y, Viegas-Pequignot E, Skorecki K, Selig S. Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum Mol Genet. 2008;17(18):2776–89.
Brun ME, Lana E, Rivals I, Lefranc G, Sarda P, Claustres M, et al. Heterochromatic genes undergo epigenetic changes and escape silencing in immunodeficiency, centromeric instability, facial anomalies (ICF) syndrome. PLoS ONE. 2011;6(4):e19464.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5.
Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 2011;333(6047):1300–3.
Hu L, Lu J, Cheng J, Rao Q, Li Z, Hou H, et al. Structural insight into substrate preference for TET-mediated oxidation. Nature. 2015;527(7576):118–22.
Hu L, Li Z, Cheng J, Rao Q, Gong W, Liu M, et al. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell. 2013;155(7):1545–55.
Lorsbach RB, Moore J, Mathew S, Raimondi SC, Mukatira ST, Downing JR. TET1, a member of a novel protein family, is fused to MLL in acute myeloid leukemia containing the t(10;11)(q22;q23). Leukemia. 2003;17(3):637–41.
Huang H, Jiang X, Li Z, Li Y, Song CX, He C, et al. TET1 plays an essential oncogenic role in MLL-rearranged leukemia. Proc Natl Acad Sci U S A. 2013;110(29):11994–9.
Good CR, Panjarian S, Kelly AD, Madzo J, Patel B, Jelinek J, et al. TET1-mediated hypomethylation activates oncogenic signaling in triple-negative breast cancer. Cancer Res. 2018;78(15):4126–37.
Zhao M, Wang J, Liao W, Li D, Li M, Wu H, et al. Increased 5-hydroxymethylcytosine in CD4(+) T cells in systemic lupus erythematosus. J Autoimmun. 2016;69:64–73.
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–50.
Guillamot M, Cimmino L, Aifantis I. The Impact of DNA methylation in hematopoietic malignancies. Trends Cancer. 2016;2(2):70–83.
Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med. 2009;360(22):2289–301.
Kosmider O, Gelsi-Boyer V, Ciudad M, Racoeur C, Jooste V, Vey N, et al. TET2 gene mutation is a frequent and adverse event in chronic myelomonocytic leukemia. Haematologica. 2009;94(12):1676–81.
Schaub FX, Looser R, Li S, Hao-Shen H, Lehmann T, Tichelli A, et al. Clonal analysis of TET2 and JAK2 mutations suggests that TET2 can be a late event in the progression of myeloproliferative neoplasms. Blood. 2010;115(10):2003–7.
Kaasinen E, Kuismin O, Rajamaki K, Ristolainen H, Aavikko M, Kondelin J, et al. Impact of constitutional TET2 haploinsufficiency on molecular and clinical phenotype in humans. Nat Commun. 2019;10(1):1252.
Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 function blocks aberrant self-renewal and leukemia progression. Cell. 2017;170(6):1079-95.e20.
Duployez N, Goursaud L, Fenwarth L, Bories C, Marceau-Renaut A, Boyer T, et al. Familial myeloid malignancies with germline TET2 mutation. Leukemia. 2020;34(5):1450–3.
•• Stremenova Spegarova J, Lawless D, Mohamad SMB, Engelhardt KR, Doody G, Shrimpton J, et al. Germline TET2 loss of function causes childhood immunodeficiency and lymphoma. Blood. 2020;136(9):1055–66. Reports on rare TET2 germline variants with a predisposition to lymphoma.
Cargo C, Cullen M, Taylor J, Short M, Glover P, Van Hoppe S, et al. The use of targeted sequencing and flow cytometry to identify patients with a clinically significant monocytosis. Blood. 2019;133(12):1325–34.
•• Beck DB, Petracovici A, He C, Moore HW, Louie RJ, Ansar M, et al. Delineation of a human Mendelian disorder of the DNA demethylation machinery: TET3 deficiency. Am J Hum Genet. 2020;106(2):234–45. First report of rare TET3 germline variants.
Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403(6769):501–2.
Iqbal K, Jin SG, Pfeifer GP, Szabo PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108(9):3642–7.
Santos-Cortez RLP, Khan V, Khan FS, Mughal ZU, Chakchouk I, Lee K, et al. Novel candidate genes and variants underlying autosomal recessive neurodevelopmental disorders with intellectual disability. Hum Genet. 2018;137(9):735–52.
Bergaggio E, Piva R. Wild-type IDH enzymes as actionable targets for cancer therapy. Cancers (Basel). 2019;11(4):563. https://doi.org/10.3390/cancers11040563.
Sciacovelli M, Schmidt C, Maher ER, Frezza C. Metabolic drivers in hereditary cancer syndromes. Ann Rev Cancer Biol. 2020;4(1):77–97.
Kranendijk M, Struys EA, van Schaftingen E, Gibson KM, Kanhai WA, van der Knaap MS, et al. IDH2 mutations in patients with D-2-hydroxyglutaric aciduria. Science. 2010;330(6002):336.
Hamadou WS, Bourdon V, Letard S, Brenet F, Laarif S, Besbes S, et al. Familial hematological malignancies: new IDH2 mutation. Ann Hematol. 2016;95(12):1943–7.
Molenaar R, Sanikommu SR, Patel BJ, Przychodzen B, van Noorden CJ, Radivoyevitch T, et al. Whole-exome sequencing identifies germline IDH2 and IDH3 mutations that predispose to myeloid neoplasms. Blood. 2015;126(23):1405-.
Bak A, Skonieczka K, Jaskowiec A, Junkiert-Czarnecka A, Heise M, Pilarska-Deltow M, et al. Germline mutations among Polish patients with acute myeloid leukemia. Hered Cancer Clin Pract. 2021;19(1):42.
Kendroud S, Groepper D, Choi YJ. Apparent germline IDH1 mutation in a patient with Ollier disease and glioblastomas: a case report (P1.9–042). Neurology. 2019;92(15 Supplement):P1.9–042.
Blackburn PR, Carter JM, Oglesbee D, Westendorf JJ, Neff BA, Stichel D, et al. An activating germline IDH1 variant associated with a tumor entity characterized by unilateral and bilateral chondrosarcoma of the mastoid. HGG Adv. 2020;1(1):100006.
Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH, et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell. 2012;22(2):180–93.
Rahmani NE, Ramachandra N, Bhagat TD, Gordon S, Pradhan K, Rivera Pena B, et al. ASXL1 mutations are associated with widespread and distinct DNA methylation alterations. Blood. 2019;134(Supplement_1):2989-.
Boultwood J, Perry J, Pellagatti A, Fernandez-Mercado M, Fernandez-Santamaria C, Calasanz MJ, et al. Frequent mutation of the polycomb-associated gene ASXL1 in the myelodysplastic syndromes and in acute myeloid leukemia. Leukemia. 2010;24(5):1062–5.
Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N, et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol. 2009;145(6):788–800.
Genovese G, Kahler AK, Handsaker RE, Lindberg J, Rose SA, Bakhoum SF, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477–87.
• Bohring A, Silengo M, Lerone M, Superneau DW, Spaich C, Braddock SR, et al. Severe end of Opitz trigonocephaly (C) syndrome or new syndrome? Am J Med Genet. 1999;85(5):438–46. Initial report of Bohring-Opitz syndrome.
Hoischen A, van Bon BWM, Rodríguez-Santiago B, Gilissen C, Vissers LELM, de Vries P, et al. De novo nonsense mutations in ASXL1 cause Bohring-Opitz syndrome. Nat Genet. 2011;43(8):729–31.
Oberklaid F, Danks DM. The Opitz trigonocephaly syndrome: a case report. Am J Dis Child. 1975;129(11):1348–9.
Hastings R, Cobben J-M, Gillessen-Kaesbach G, Goodship J, Hove H, Kjaergaard S, et al. Bohring-Opitz (Oberklaid–Danks) syndrome: clinical study, review of the literature, and discussion of possible pathogenesis. Eur J Hum Genet. 2011;19(5):513–9.
Bohring A, Oudesluijs GG, Grange DK, Zampino G, Thierry P. New cases of Bohring-Opitz syndrome, update, and critical review of the literature. Am J Med Genet A. 2006;140A(12):1257–63.
Micol JB, Abdel-Wahab O. The role of additional sex combs-like proteins in cancer. Cold Spring Harb Perspect Med. 2016;6(10):a026526. https://doi.org/10.1101/cshperspect.a026526.
Russell B, Johnston JJ, Biesecker LG, Kramer N, Pickart A, Rhead W, et al. Clinical management of patients with ASXL1 mutations and Bohring-Opitz syndrome, emphasizing the need for Wilms tumor surveillance. Am J Med Genet A. 2015;167A(9):2122–31.
Seiter K, Htun K, Baskind P, Liu Z. Acute myeloid leukemia in a father and son with a germline mutation of ASXL1. Biomark Res. 2018;6:7.
Hamadou WS, Abed RE, Besbes S, Bourdon V, Fabre A, Youssef YB, et al. Familial hematological malignancies: ASXL1 gene investigation. Clin Transl Oncol. 2016;18(4):385–90.
Dinwiddie DL, Soden SE, Saunders CJ, Miller NA, Farrow EG, Smith LD, et al. De novo frameshift mutation in ASXL3 in a patient with global developmental delay, microcephaly, and craniofacial anomalies. BMC Med Genomics. 2013;6:32.
Bainbridge MN, Hu H, Muzny DM, Musante L, Lupski JR, Graham BH, et al. De novo truncating mutations in ASXL3 are associated with a novel clinical phenotype with similarities to Bohring-Opitz syndrome. Genome Med. 2013;5(2):11.
Yang L, Guo B, Zhu W, Wang L, Han B, Che Y, et al. Bainbridge-ropers syndrome caused by loss-of-function variants in ASXL3: clinical abnormalities, medical imaging features, and gene variation in infancy of case report. BMC Pediatr. 2020;20(1):287.
Shashi V, Pena LD, Kim K, Burton B, Hempel M, Schoch K, et al. De novo truncating variants in ASXL2 are associated with a unique and recognizable clinical phenotype. Am J Hum Genet. 2016;99(4):991–9.
Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16(22):2893–905.
Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–43.
Vire E, Brenner C, Deplus R, Blanchon L, Fraga M, Didelot C, et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439(7078):871–4.
Guan X, Deng H, Choi UL, Li Z, Yang Y, Zeng J, et al. EZH2 overexpression dampens tumor-suppressive signals via an EGR1 silencer to drive breast tumorigenesis. Oncogene. 2020;39(48):7127–41.
Ernst T, Chase AJ, Score J, Hidalgo-Curtis CE, Bryant C, Jones AV, et al. Inactivating mutations of the histone methyltransferase gene EZH2 in myeloid disorders. Nat Genet. 2010;42(8):722–6.
Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G, et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med. 2011;364(26):2496–506.
• Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, et al. Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90(1):110–8. Identifies EZH2 mutations as the cause of Weaver syndrome.
Weaver DD, Graham CB, Thomas IT, Smith DW. A new overgrowth syndrome with accelerated skeletal maturation, unusual facies, and camptodactyly. J Pediatr. 1974;84(4):547–52.
Tatton-Brown K, Murray A, Hanks S, Douglas J, Armstrong R, Banka S, et al. Weaver syndrome and EZH2 mutations: clarifying the clinical phenotype. Am J Med Genet A. 2013;161A(12):2972–80.
Cohen AS, Yap DB, Lewis ME, Chijiwa C, Ramos-Arroyo MA, Tkachenko N, et al. Weaver syndrome-associated EZH2 protein variants show impaired histone methyltransferase function in vitro. Hum Mutat. 2016;37(3):301–7.
•• Basel-Vanagaite L. Acute lymphoblastic leukemia in Weaver syndrome. Am J Med Genet A. 2010;152A(2):383–6. Reports on the predisposition to malignancy in Weaver syndrome.
Funding
Mrinal Patnaik has received research funding from Stem Line Pharmaceuticals and Kura Oncology.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Jenna Fernandez declares no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Germline Predisposition to Myeloid Neoplasms
Rights and permissions
About this article
Cite this article
Fernandez, J.A., Patnaik, M.M. Germline Abnormalities in DNA Methylation and Histone Modification and Associated Cancer Risk. Curr Hematol Malig Rep 17, 82–93 (2022). https://doi.org/10.1007/s11899-022-00665-5
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-022-00665-5