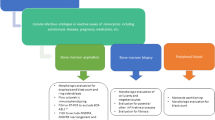
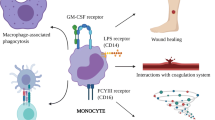
Abstract
Purpose of Review
In this review, we provide a comprehensive and contemporary understanding of malignant monocytosis and provide a framework by which the appropriate diagnosis with malignant monocytosis can be rendered.
Recent Findings
Increasing data support the use of molecular data to refine the diagnostic approach to persistent monocytosis. The absence of a TET2, SRSF2, or ASXL1 mutation has ≥ 90% negative predictive value for a diagnosis of CMML. These data may also reliably differentiate chronic myelomonocytic leukemia, the malignancy that is most associated with mature monocytosis, from several other diseases that can be associated with typically a lesser degree of monocytosis. These include acute myelomonocytic leukemia, acute myeloid leukemia with monocytic differentiation, myelodysplastic syndromes, and myeloproliferative neoplasms driven by BCR-ABL1, PDGFRA, PDGFRB, or FGFR1 rearrangements or PCM1-JAK2 fusions among other rarer aberrations. The combination of monocyte partitioning with molecular data in patients with persistent monocytosis may increase the predictive power for the ultimate development of CMM but has not been prospectively validated.
Summary
Many conditions, both benign and malignant, can be associated with an increase in mature circulating monocytes. After reasonably excluding a secondary or reactive monocytosis, there should be a concern for and investigation of malignant monocytosis, which includes hematopathologic review of blood and marrow tissues, flow cytometric analysis, and cytogenetic and molecular studies to arrive at an appropriate diagnosis.





Similar content being viewed by others
References
Hettinger J, Richards DM, Hansson J, Barra MM, Joschko AC, Krijgsveld J, et al. Origin of monocytes and macrophages in a committed progenitor. Nat Immunol. 2013;14:821–30.
Auffray C, Sieweke MH, Geissmann F. Blood monocytes: development, heterogeneity, and relationship with dendritic cells. Annu Rev Immunol. 2009;27:669–92.
Ziegler-Heitbrock L, Ancuta P, Crowe S, Dalod M, Grau V, Hart DN, et al. Nomenclature of monocytes and dendritic cells in blood. Blood. 2010;116:e74–80.
Rees AJ. Monocyte and macrophage biology: an overview. Semin Nephrol. 2010;30:216–33.
Ley K, Laudanna C, Cybulsky MI, Nourshargh S. Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat Rev Immunol. 2007;7:678–89.
Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44:450–62.
Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, et al., editors. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC; 2017.
Ahmed SG, Bukar AA, Jolayemi B. Hematological indices of sickle cell anaemia patients with pulmonary tuberculosis in northern Nigeria. Mediterr J Hematol Infect Dis. 2010;2:e2010014.
Kini RG, Chandrashekhar J. Parasite and the circulating pool-characterisation of leukocyte number and morphology in malaria. J Clin Diagn Res. 2016;10:EC44–8.
Bardina SV, Michlmayr D, Hoffman KW, Obara CJ, Sum J, Charo IF, et al. Differential roles of chemokines CCL2 and CCL7 in monocytosis and leukocyte migration during West Nile virus infection. J Immunol. 2015;195:4306–18.
Hensel M, Gradel L, Kutz A, Haubitz S, Huber A, Mueller B, et al. Peripheral monocytosis as a predictive factor for adverse outcome in the emergency department: survey based on a register study. Medicine (Baltimore). 2017;96:e7404.
Thone J, Kessler E. Monocytosis subsequent to ziprasidone treatment: a possible side effect. Prim Care Companion J Clin Psychiatry. 2007;9:465–6.
Garman L, Pelikan RC, Rasmussen A, Lareau CA, Savoy KA, Deshmukh US, et al. Single cell transcriptomics implicate novel monocyte and T cell immune dysregulation in sarcoidosis. Front Immunol. 2020;11:567342.
Wurm K, Lohr G. Immuno-cytological blood tests in cases of sarcoidosis. Sarcoidosis. 1986;3:52–9.
Sumegi A, Antal-Szalmas P, Aleksza M, Kovacs I, Sipka S, Zeher M, et al. Glucocorticosteroid therapy decreases CD14-expression and CD14-mediated LPS-binding and activation of monocytes in patients suffering from systemic lupus erythematosus. Clin Immunol. 2005;117:271–9.
Lescoat A, Lecureur V, Roussel M, Sunnaram BL, Ballerie A, Coiffier G, et al. CD16-positive circulating monocytes and fibrotic manifestations of systemic sclerosis. Clin Rheumatol. 2017;36:1649–54.
Higashi-Kuwata N, Jinnin M, Makino T, Fukushima S, Inoue Y, Muchemwa FC, et al. Characterization of monocyte/macrophage subsets in the skin and peripheral blood derived from patients with systemic sclerosis. Arthritis Res Ther. 2010;12:R128.
Demeter J, Mihalik R, Benczur M, Lehoczky D, Paloczi K. Peripheral blood leukocyte subpopulations a long time after posttraumatic splenectomy. Haematologia (Budap). 1991;24:139–44.
Grabowski S. A case of monocytosis after splenectomy. Pol Tyg Lek (Wars). 1952;7:795–6.
Swirski FK, Nahrendorf M, Etzrodt M, Wildgruber M, Cortez-Retamozo V, Panizzi P, et al. Identification of splenic reservoir monocytes and their deployment to inflammatory sites. Science. 2009;325:612–6.
Bhatnagar S, Chandra J, Narayan S. Hematological changes and predictors of bone marrow recovery in patients with neutropenic episodes in acute lymphoblastic leukemia. J Trop Pediatr. 2002;48:200–3.
Le Bourgeois A, Lestang E, Guillaume T, Delaunay J, Ayari S, Blin N, et al. Prognostic impact of immune status and hematopoietic recovery before and after fludarabine, IV busulfan, and antithymocyte globulins (FB2 regimen) reduced-intensity conditioning regimen (RIC) allogeneic stem cell transplantation (allo-SCT). Eur J Haematol. 2013;90:177–86.
Nishiwaki U, Nakayama S, Yokote T, Hiraoka N, Tsuji M. Classical Hodgkin lymphoma producing macrophage colony-stimulating factor with resultant monocytosis. Br J Haematol. 2017;176:343.
Tadmor T, Fell R, Polliack A, Attias D. Absolute monocytosis at diagnosis correlates with survival in diffuse large B-cell lymphoma-possible link with monocytic myeloid-derived suppressor cells. Hematol Oncol. 2013;31:65–71.
Maran R, Mittelman M, Cohen AM, Djaldetti M. Myelokathexis and monocytosis in a patient with gastric cancer. Acta Haematol. 1992;87:210–2.
Eo WK, Kwon BS, Kim KH, Kim HY, Kim HB, Koh SB, et al. Monocytosis as a prognostic factor for survival in stage IB and IIA cervical cancer. J Cancer. 2018;9:64–70.
Patnaik MM, Tefferi A. Chronic myelomonocytic leukemia: 2018 update on diagnosis, risk stratification and management. Am J Hematol. 2018;93:824–40.
Kerrigan DP, Castillo A, Foucar K, Townsend K, Neidhart J. Peripheral blood morphologic changes after high-dose antineoplastic chemotherapy and recombinant human granulocyte colony-stimulating factor administration. Am J Clin Pathol. 1989;92:280–5.
Lynch DT, Hall J, Foucar K. How I investigate monocytosis. Int J Lab Hematol. 2018;40:107–14.
Xu Y, McKenna RW, Karandikar NJ, Pildain AJ, Kroft SH. Flow cytometric analysis of monocytes as a tool for distinguishing chronic myelomonocytic leukemia from reactive monocytosis. Am J Clin Pathol. 2005;124:799–806.
Lacronique-Gazaille C, Chaury MP, Le Guyader A, Faucher JL, Bordessoule D, Feuillard J. A simple method for detection of major phenotypic abnormalities in myelodysplastic syndromes: expression of CD56 in CMML. Haematologica. 2007;92:859–60.
Selimoglu-Buet D, Wagner-Ballon O, Saada V, Bardet V, Itzykson R, Bencheikh L, et al. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood. 2015;125:3618–26.
Goasguen JE, Bennett JM, Bain BJ, Vallespi T, Brunning R, Mufti GJ, et al. Morphological evaluation of monocytes and their precursors. Haematologica. 2009;94:994–7.
Yang DT, Greenwood JH, Hartung L, Hill S, Perkins SL, Bahler DW. Flow cytometric analysis of different CD14 epitopes can help identify immature monocytic populations. Am J Clin Pathol. 2005;124:930–6.
Foucar K, Hsi ED, Wang SA, Rogers HJ, Hasserjian RP, Bagg A, et al. Concordance among hematopathologists in classifying blasts plus promonocytes: a bone marrow pathology group study. Int J Lab Hematol. 2020;42:418–22.
Shallis RM, Xu ML, Podoltsev NA, Curtis SA, Considine BT, Khanna SR, et al. Be careful of the masquerades: differentiating secondary myelodysplasia from myelodysplastic syndromes in clinical practice. Ann Hematol. 2018;97:2333–43.
Mason CC, Khorashad JS, Tantravahi SK, Kelley TW, Zabriskie MS, Yan D, et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia. 2016;30:906–13.
Palomo L, Garcia O, Arnan M, Xicoy B, Fuster F, Cabezon M, et al. Targeted deep sequencing improves outcome stratification in chronic myelomonocytic leukemia with low risk cytogenetic features. Oncotarget. 2016;7:57021–35.
Hwang SM, Kim SM, Nam Y, Kim J, Kim S, Ahn YO, et al. Targeted sequencing aids in identifying clonality in chronic myelomonocytic leukemia. Leuk Res. 2019;84:106190.
Kohlmann A, Grossmann V, Klein HU, Schindela S, Weiss T, Kazak B, et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J Clin Oncol. 2010;28:3858–65.
Meggendorfer M, Roller A, Haferlach T, Eder C, Dicker F, Grossmann V, et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood. 2012;120:3080–8.
Itzykson R, Kosmider O, Renneville A, Morabito M, Preudhomme C, Berthon C, et al. Clonal architecture of chronic myelomonocytic leukemias. Blood. 2013;121:2186–98.
Itzykson R, Solary E. An evolutionary perspective on chronic myelomonocytic leukemia. Leukemia. 2013;27:1441–50.
Patnaik MM, Itzykson R, Lasho TL, Kosmider O, Finke CM, Hanson CA, et al. ASXL1 and SETBP1 mutations and their prognostic contribution in chronic myelomonocytic leukemia: a two-center study of 466 patients. Leukemia. 2014;28:2206–12.
Merlevede J, Droin N, Qin T, Meldi K, Yoshida K, Morabito M, et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat Commun. 2016;7:10767.
Shallis RM, Ahmad R, Zeidan AM. The genetic and molecular pathogenesis of myelodysplastic syndromes. Eur J Haematol. 2018;101:260–71.
Patnaik MM, Parikh SA, Hanson CA, Tefferi A. Chronic myelomonocytic leukaemia: a concise clinical and pathophysiological review. Br J Haematol. 2014;165:273–86.
Patnaik MM, Tefferi A. Cytogenetic and molecular abnormalities in chronic myelomonocytic leukemia. Blood Cancer J. 2016;6:e393.
Cargo C, Cullen M, Taylor J, Short M, Glover P, Van Hoppe S, et al. The use of targeted sequencing and flow cytometry to identify patients with a clinically significant monocytosis. Blood. 2019;133:1325–34.
Schuler E, Frank F, Hildebrandt B, Betz B, Strupp C, Rudelius M, et al. Myelodysplastic syndromes without peripheral monocytosis but with evidence of marrow monocytosis share clinical and molecular characteristics with CMML. Leuk Res. 2018;65:1–4.
Selimoglu-Buet D, Badaoui B, Benayoun E, Toma A, Fenaux P, Quesnel B, et al. Accumulation of classical monocytes defines a subgroup of MDS that frequently evolves into CMML. Blood. 2017;130:832–5.
Talati C, Zhang L, Shaheen G, Kuykendall A, Ball M, Zhang Q, et al. Monocyte subset analysis accurately distinguishes CMML from MDS and is associated with a favorable MDS prognosis. Blood. 2017;129:1881–3.
Meggendorfer M, Jeromin S, Haferlach C, Kern W, Haferlach T. The mutational landscape of 18 investigated genes clearly separates four subtypes of myelodysplastic/myeloproliferative neoplasms. Haematologica. 2018;103:e192–e5.
Barraco D, Cerquozzi S, Gangat N, Patnaik MM, Lasho T, Finke C, et al. Monocytosis in polycythemia vera: Clinical and molecular correlates. Am J Hematol. 2017;92:640–5.
Elliott MA, Verstovsek S, Dingli D, Schwager SM, Mesa RA, Li CY, et al. Monocytosis is an adverse prognostic factor for survival in younger patients with primary myelofibrosis. Leuk Res. 2007;31:1503–9.
Boiocchi L, Espinal-Witter R, Geyer JT, Steinhilber J, Bonzheim I, Knowles DM, et al. Development of monocytosis in patients with primary myelofibrosis indicates an accelerated phase of the disease. Mod Pathol. 2013;26:204–12.
Wang SA, Tam W, Tsai AG, Arber DA, Hasserjian RP, Geyer JT, et al. Targeted next-generation sequencing identifies a subset of idiopathic hypereosinophilic syndrome with features similar to chronic eosinophilic leukemia, not otherwise specified. Mod Pathol. 2016;29:854–64.
Bennett JM, Catovsky D, Daniel MT, Flandrin G, Galton DA, Gralnick H, et al. The chronic myeloid leukaemias: guidelines for distinguishing chronic granulocytic, atypical chronic myeloid, and chronic myelomonocytic leukaemia. Proposals by the French-American-British Cooperative Leukaemia Group. Br J Haematol. 1994;87:746–54.
Wang X, Wang F, Wang Z, Li Y, Wang D, Wu H, et al. p210(BCR-ABL1)- chronic myeloid leukemia presents with monocytosis. Clin Case Rep. 2020;8:840–2.
Gatti A, Movilia A, Roncoroni L, Citro A, Marinoni S, Brando B. Chronic myeloid leukemia with P190 BCR-ABL translocation and persistent moderate monocytosis: a case report. J Hematol. 2018;7:120–3.
Dass J, Jain S, Tyagi S, Sazawal S. Chronic myeloid leukemia with p210 BCR-ABL and monocytosis. Leuk Lymphoma. 2011;52:1380–1.
Patnaik MM, Timm MM, Vallapureddy R, Lasho TL, Ketterling RP, Gangat N, et al. Flow cytometry based monocyte subset analysis accurately distinguishes chronic myelomonocytic leukemia from myeloproliferative neoplasms with associated monocytosis. Blood Cancer J. 2017;7:e584.
Patnaik MM, Lasho TL, Finke CM, Pardanani A, Tefferi A. Targeted next generation sequencing of PDGFRB rearranged myeloid neoplasms with monocytosis. Am J Hematol. 2016;91:E12–4.
Macdonald D, Reiter A, Cross NC. The 8p11 myeloproliferative syndrome: a distinct clinical entity caused by constitutive activation of FGFR1. Acta Haematol. 2002;107:101–7.
Liu Y, Mi X, Gadde R, Gao Q, Xiao W, Zhang Y, et al. FGFR1 Rearrangement guides diagnosis and treatment of a trilineage B, T, and myeloid mixed phenotype acute leukemia. JCO Precis Oncol. 2020;4.
Kumar KR, Chen W, Koduru PR, Luu HS. Myeloid and lymphoid neoplasm with abnormalities of FGFR1 presenting with trilineage blasts and RUNX1 rearrangement: a case report and review of literature. Am J Clin Pathol. 2015;143:738–48.
Kasbekar M, Nardi V, Dal Cin P, Brunner AM, Burke M, Chen YB, et al. Targeted FGFR inhibition results in a durable remission in an FGFR1-driven myeloid neoplasm with eosinophilia. Blood Adv. 2020;4:3136–40.
Bain BJ, Ahmad S. Should myeloid and lymphoid neoplasms with PCM1-JAK2 and other rearrangements of JAK2 be recognized as specific entities? Br J Haematol. 2014;166:809–17.
Apperley JF, Gardembas M, Melo JV, Russell-Jones R, Bain BJ, Baxter EJ, et al. Response to imatinib mesylate in patients with chronic myeloproliferative diseases with rearrangements of the platelet-derived growth factor receptor beta. N Engl J Med. 2002;347:481–7.
Bain BJ. Myeloid and lymphoid neoplasms with eosinophilia and abnormalities of PDGFRA, PDGFRB or FGFR1. Haematologica. 2010;95:696–8.
Metzgeroth G, Schwaab J, Gosenca D, Fabarius A, Haferlach C, Hochhaus A, et al. Long-term follow-up of treatment with imatinib in eosinophilia-associated myeloid/lymphoid neoplasms with PDGFR rearrangements in blast phase. Leukemia. 2013;27:2254–6.
Meggendorfer M, Haferlach C, Zenger M, Macijewski K, Kern W, Haferlach T. The landscape of myeloid neoplasms with isochromosome 17q discloses a specific mutation profile and is characterized by an accumulation of prognostically adverse molecular markers. Leukemia. 2016;30:1624–7.
McClure RF, Dewald GW, Hoyer JD, Hanson CA. Isolated isochromosome 17q: a distinct type of mixed myeloproliferative disorder/myelodysplastic syndrome with an aggressive clinical course. Br J Haematol. 1999;106:445–54.
Kanagal-Shamanna R, Bueso-Ramos CE, Barkoh B, Lu G, Wang S, Garcia-Manero G, et al. Myeloid neoplasms with isolated isochromosome 17q represent a clinicopathologic entity associated with myelodysplastic/myeloproliferative features, a high risk of leukemic transformation, and wild-type TP53. Cancer. 2012;118:2879–88.
Kanagal-Shamanna R, Luthra R, Yin CC, Patel KP, Takahashi K, Lu X, et al. Myeloid neoplasms with isolated isochromosome 17q demonstrate a high frequency of mutations in SETBP1, SRSF2, ASXL1 and NRAS. Oncotarget. 2016;7:14251–8.
Code Availability
Not applicable.
Availability of Data and Material
Not applicable.
Author information
Authors and Affiliations
Contributions
R.M.S. and A.M.Z. conceptualized and wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
A.M.Z. received research funding (institutional) from Celgene, Acceleron, Abbvie, Otsuka, Pfizer, Medimmune/AstraZeneca, Boehringer-Ingelheim, Trovagene, Incyte, Takeda, and ADC Therapeutics. A.M.Z had a consultancy with and received honoraria from AbbVie, Otsuka, Pfizer, Celgene, Ariad, Incyte, Agios, Boehringer-Ingelheim, Novartis, Acceleron, Astellas, Daiichi Sankyo, Cardinal Health, Seattle Genetics, BeyondSpring, and Takeda. None of these relationships were related to the development of this manuscript. All other authors report no relevant disclosures/competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Myelodysplastic Syndromes and MPN/MDS Overlap
Rights and permissions
About this article
Cite this article
Shallis, R.M., Siddon, A.J. & Zeidan, A.M. Clinical and Molecular Approach to Adult-Onset, Neoplastic Monocytosis. Curr Hematol Malig Rep 16, 276–285 (2021). https://doi.org/10.1007/s11899-021-00632-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-021-00632-6