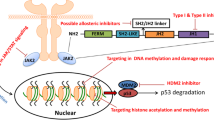
Abstract
Purpose of Review
T cell lymphoproliferative disorders represent a diverse group of hematologic malignancies with poor prognosis underscoring the need for novel therapeutic approaches. Disruption of the JAK/STAT signaling pathway has been described in this group of blood cancers and may represent an approach for targeted therapy. Here, we summarize the current data describing the disruptions of JAK/STAT signaling in T cell malignancies and focus on the existing evidence for exploitation of this pathway with targeted therapies.
Recent Findings
To date, preclinical studies have demonstrated the efficacy of JAK/STAT inhibition in the treatment of several T cell lymphoproliferative disorders. More recently, several early clinical trials have demonstrated promising results utilizing this approach as well. The benefit of the combination of JAK/STAT-targeted therapies along with immunotherapy and other molecularly targeted therapies is also discussed.
Summary
There is substantial evidence that targeting the JAK/STAT pathway in T cell lymphoproliferative disorders could be of clinical benefit. There are several early clinical trials showing promise and many ongoing trials investigating the optimal utility of agents that inhibit this signaling pathway. In addition, targeting this pathway may provide a platform for further rational combination therapies.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
O’Shea JJ, Schwartz DM, Villarino AV, Gadina M, McInnes IB, Laurence A. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–28.
Springuel L, Renauld JC, Knoops L. JAK kinase targeting in hematologic malignancies: a sinuous pathway from identification of genetic alterations towards clinical indications. Haematologica. 2015;100(10):1240–53.
Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004;117(Pt 8):1281–3.
James C, Ugo V, Le Couedic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature. 2005;434(7037):1144–8.
Pencik J, Pham HT, Schmoellerl J, Javaheri T, Schlederer M, Culig Z, et al. JAK-STAT signaling in cancer: from cytokines to non-coding genome. Cytokine. 2016;87:26–36.
Groner B, von Manstein V. JAK STAT signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14.
Seif F, Khoshmirsafa M, Aazami H, Mohsenzadegan M, Sedighi G, Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun Signal. 2017;15(1):23.
Matutes E. The 2017 WHO update on mature T- and natural killer (NK) cell neoplasms. Int J Lab Hematol. 2018;40(Suppl 1):97–103.
Finbloom DS, Larner AC. Regulation of the JAK/STAT signalling pathway. Cell Signal. 1995;7(8):739–45.
Vainchenker W, Leroy E, Gilles L, Marty C, Plo I, Constantinescu SN. JAK inhibitors for the treatment of myeloproliferative neoplasms and other disorders. F1000Res. 2018;7:82.
Zhang Y, Li C, Xue W, Zhang M, Li Z. Frequent mutations in natural killer/T cell lymphoma. Cell Physiol Biochem. 2018;49(1):1–16.
Dobashi A, Tsuyama N, Asaka R, Togashi Y, Ueda K, Sakata S, et al. Frequent BCOR aberrations in extranodal NK/T-cell lymphoma, nasal type. Genes Chromosom Cancer. 2016;55(5):460–71.
Kucuk C, Jiang B, Hu X, Zhang W, Chan JK, Xiao W, et al. Activating mutations of STAT5B and STAT3 in lymphomas derived from gammadelta-T or NK cells. Nat Commun. 2015;6:6025.
Jiang L, Gu ZH, Yan ZX, Zhao X, Xie YY, Zhang ZG, et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat Genet. 2015;47(9):1061–6.
Lee S, Park HY, Kang SY, Kim SJ, Hwang J, Lee S, et al. Genetic alterations of JAK/STAT cascade and histone modification in extranodal NK/T-cell lymphoma nasal type. Oncotarget. 2015;6(19):17764–76.
Koo GC, Tan SY, Tang T, Poon SL, Allen GE, Tan L, et al. Janus kinase 3-activating mutations identified in natural killer/T-cell lymphoma. Cancer Discov. 2012;2(7):591–7.
Bouchekioua A, Scourzic L, de Wever O, Zhang Y, Cervera P, Aline-Fardin A, et al. JAK3 deregulation by activating mutations confers invasive growth advantage in extranodal nasal-type natural killer cell lymphoma. Leukemia. 2014;28(2):338–48.
Kimura H, Karube K, Ito Y, Hirano K, Suzuki M, Iwata S, et al. Rare occurrence of JAK3 mutations in natural killer cell neoplasms in Japan. Leuk Lymphoma. 2014;55(4):962–3.
Sim SH, Kim S, Kim TM, Jeon YK, Nam SJ, Ahn YO, et al. Novel JAK3-activating mutations in extranodal NK/T-cell lymphoma. Nasal Type Am J Pathol. 2017;187(5):980–6.
Chen YW, Guo T, Shen L, Wong KY, Tao Q, Choi WW, et al. Receptor-type tyrosine-protein phosphatase kappa directly targets STAT3 activation for tumor suppression in nasal NK/T-cell lymphoma. Blood. 2015;125(10):1589–600.
de Mel S, Soon GST, Mok Y, Chung TH, Jeyasekharan AD, Chng WJ, et al. The genomics and molecular biology of natural killer/T-cell lymphoma: opportunities for translation. Int J Mol Sci. 2018;19(7):1931.
Song TL, Nairismagi ML, Laurensia Y, Lim JQ, Tan J, Li ZM, et al. Oncogenic activation of the STAT3 pathway drives PD-L1 expression in natural killer/T-cell lymphoma. Blood. 2018;132(11):1146–58.
Hofmann SR, Lam AQ, Frank S, Zhou YJ, Ramos HL, Kanno Y, et al. Jak3-independent trafficking of the common gamma chain receptor subunit: chaperone function of Jaks revisited. Mol Cell Biol. 2004;24(11):5039–49.
Johnston JA, Kawamura M, Kirken RA, Chen YQ, Blake TB, Shibuya K, et al. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. 1994;370(6485):151–3.
•• Nairismagi M, Gerritsen ME, Li ZM, Wijaya GC, Chia BKH, Laurensia Y, et al. Oncogenic activation of JAK3-STAT signaling confers clinical sensitivity to PRN371, a novel selective and potent JAK3 inhibitor, in natural killer/T-cell lymphoma. Leukemia. 2018;32(5):1147–56 The investigators demonstrate the efficacy of specific JAK3 inhibition thereby overcoming a potential limitation of off-target effects of prior pan-JAK inhibitors.
•• Kwong YL, Chan TSY, Tan D, Kim SJ, Poon LM, Mow B, et al. PD1 blockade with pembrolizumab is highly effective in relapsed or refractory NK/T-cell lymphoma failing l-asparaginase. Blood. 2017;129(17):2437–42 Demonstration of the efficacy of PD1-based immunotherapy in treating NK/T cell lymphoma.
• Dufva O, Kankainen M, Kelkka T, Sekiguchi N, Awad SA, Eldfors S, et al. Aggressive natural killer-cell leukemia mutational landscape and drug profiling highlight JAK-STAT signaling as therapeutic target. Nat Commun. 2018;9(1):1567 Demonstration of JAK-STAT signaling as a therapeutic target in aggressive forms of NK cell leukemia, a disease with very poor outcomes.
Jerez A, Clemente MJ, Makishima H, Koskela H, Leblanc F, Peng Ng K, et al. STAT3 mutations unify the pathogenesis of chronic lymphoproliferative disorders of NK cells and T-cell large granular lymphocyte leukemia. Blood. 2012;120(15):3048–57.
Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–13.
Rajala HL, Eldfors S, Kuusanmaki H, van Adrichem AJ, Olson T, Lagstrom S, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–50.
Rajala HL, Mustjoki S. STAT5b in LGL leukemia--a novel therapeutic target? Oncotarget. 2013;4(6):808–9.
Ohgami RS, Ma L, Merker JD, Martinez B, Zehnder JL, Arber DA. STAT3 mutations are frequent in CD30+ T-cell lymphomas and T-cell large granular lymphocytic leukemia. Leukemia. 2013;27(11):2244–7.
Bamford S, Dawson E, Forbes S, Clements J, Pettett R, Dogan A, et al. The COSMIC (Catalogue of Somatic Mutations in Cancer) database and website. Br J Cancer. 2004;91(2):355–8.
Odejide O, Weigert O, Lane AA, Toscano D, Lunning MA, Kopp N, et al. A targeted mutational landscape of angioimmunoblastic T-cell lymphoma. Blood. 2014;123(9):1293–6.
Crescenzo R, Abate F, Lasorsa E, Tabbo F, Gaudiano M, Chiesa N, et al. Convergent mutations and kinase fusions lead to oncogenic STAT3 activation in anaplastic large cell lymphoma. Cancer Cell. 2015;27(4):516–32.
McKinney M, Moffitt AB, Gaulard P, Travert M, De Leval L, Nicolae A, et al. The genetic basis of hepatosplenic T-cell lymphoma. Cancer Discov. 2017;7(4):369–79.
Nicolae A, Xi L, Pittaluga S, Abdullaev Z, Pack SD, Chen J, et al. Frequent STAT5B mutations in gammadelta hepatosplenic T-cell lymphomas. Leukemia. 2014;28(11):2244–8.
• Simpson HM, Furusawa A, Sadashivaiah K, Civin CI, Banerjee A. STAT5 inhibition induces TRAIL/DR4 dependent apoptosis in peripheral T-cell lymphoma. Oncotarget. 2018;9(24):16792–806 Demonstration of the utility of STAT5 inhibition in PTCL.
Bastidas Torres AN, Najidh S, Tensen CP, Vermeer MH. Molecular advances in cutaneous T-cell lymphoma. Semin Cutan Med Surg. 2018;37(1):81–6.
Park J, Yang J, Wenzel AT, Ramachandran A, Lee WJ, Daniels JC, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E). Blood. 2017;130(12):1430–40.
da Silva Almeida AC, Abate F, Khiabanian H, Martinez-Escala E, Guitart J, Tensen CP, et al. The mutational landscape of cutaneous T cell lymphoma and Sezary syndrome. Nat Genet. 2015;47(12):1465–70.
McGirt LY, Jia P, Baerenwald DA, Duszynski RJ, Dahlman KB, Zic JA, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood. 2015;126(4):508–19.
Kricheldorff J, Fallenberg EM, Solbach C, Gerber-Schafer C, Rancso C, Fritschen UV. Breast implant-associated lymphoma. Dtsch Arztebl Int. 2018;115(38):628–35.
• Blombery P, Thompson E, Ryland GL, Joyce R, Byrne DJ, Khoo C, et al. Frequent activating STAT3 mutations and novel recurrent genomic abnormalities detected in breast implant-associated anaplastic large cell lymphoma. Oncotarget. 2018;9(90):36126–36 BIA-ALCL is shown to harbor a high frequency of STAT3 mutations.
Jeong EG, Kim MS, Nam HK, Min CK, Lee S, Chung YJ, et al. Somatic mutations of JAK1 and JAK3 in acute leukemias and solid cancers. Clin Cancer Res. 2008;14(12):3716–21.
Flex E, Petrangeli V, Stella L, Chiaretti S, Hornakova T, Knoops L, et al. Somatically acquired JAK1 mutations in adult acute lymphoblastic leukemia. J Exp Med. 2008;205(4):751–8.
Roberts KG, Morin RD, Zhang J, Hirst M, Zhao Y, Su X, et al. Genetic alterations activating kinase and cytokine receptor signaling in high-risk acute lymphoblastic leukemia. Cancer Cell. 2012;22(2):153–66.
Bains T, Heinrich MC, Loriaux MM, Beadling C, Nelson D, Warrick A, et al. Newly described activating JAK3 mutations in T-cell acute lymphoblastic leukemia. Leukemia. 2012;26(9):2144–6.
Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481(7380):157–63.
Sanda T, Tyner JW, Gutierrez A, Ngo VN, Glover J, Chang BH, et al. TYK2-STAT1-BCL2 pathway dependence in T-cell acute lymphoblastic leukemia. Cancer Discov. 2013;3(5):564–77.
Shochat C, Tal N, Bandapalli OR, Palmi C, Ganmore I, te Kronnie G, et al. Gain-of-function mutations in interleukin-7 receptor-alpha (IL7R) in childhood acute lymphoblastic leukemias. J Exp Med. 2011;208(5):901–8.
Zenatti PP, Ribeiro D, Li W, Zuurbier L, Silva MC, Paganin M, et al. Oncogenic IL7R gain-of-function mutations in childhood T-cell acute lymphoblastic leukemia. Nat Genet. 2011;43(10):932–9.
Kleppe M, Lahortiga I, El Chaar T, De Keersmaecker K, Mentens N, Graux C, et al. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat Genet. 2010;42(6):530–5.
Kleppe M, Soulier J, Asnafi V, Mentens N, Hornakova T, Knoops L, et al. PTPN2 negatively regulates oncogenic JAK1 in T-cell acute lymphoblastic leukemia. Blood. 2011;117(26):7090–8.
Porcu M, Kleppe M, Gianfelici V, Geerdens E, De Keersmaecker K, Tartaglia M, et al. Mutation of the receptor tyrosine phosphatase PTPRC (CD45) in T-cell acute lymphoblastic leukemia. Blood. 2012;119(19):4476–9.
Bellanger D, Jacquemin V, Chopin M, Pierron G, Bernard OA, Ghysdael J, et al. Recurrent JAK1 and JAK3 somatic mutations in T-cell prolymphocytic leukemia. Leukemia. 2014;28(2):417–9.
Bergmann AK, Schneppenheim S, Seifert M, Betts MJ, Haake A, Lopez C, et al. Recurrent mutation of JAK3 in T-cell prolymphocytic leukemia. Genes Chromosom Cancer. 2014;53(4):309–16.
Kiel MJ, Velusamy T, Rolland D, Sahasrabuddhe AA, Chung F, Bailey NG, et al. Integrated genomic sequencing reveals mutational landscape of T-cell prolymphocytic leukemia. Blood. 2014;124(9):1460–72.
•• Senkevitch E, Li W, Hixon JA, Andrews C, Cramer SD, Pauly GT, et al. Inhibiting Janus kinase 1 and BCL-2 to treat T cell acute lymphoblastic leukemia with IL7-Ralpha mutations. Oncotarget. 2018;9(32):22605–17 The investigators demonstrate the efficacy of combination therapy with JAK1 inhibition and BCL-2 inhibition.
Cheng Z, Yi Y, Xie S, Yu H, Peng H, Zhang G. The effect of the JAK2 inhibitor TG101209 against T cell acute lymphoblastic leukemia (T-ALL) is mediated by inhibition of JAK-STAT signaling and activation of the crosstalk between apoptosis and autophagy signaling. Oncotarget. 2017;8(63):106753–63.
Delgado-Martin C, Meyer LK, Huang BJ, Shimano KA, Zinter MS, Nguyen JV, et al. JAK/STAT pathway inhibition overcomes IL7-induced glucocorticoid resistance in a subset of human T-cell acute lymphoblastic leukemias. Leukemia. 2017;31(12):2568–76.
Waldmann TA. JAK/STAT pathway directed therapy of T-cell leukemia/lymphoma: inspired by functional and structural genomics. Mol Cell Endocrinol. 2017;451:66–70.
Zhang M, Mathews Griner LA, Ju W, Duveau DY, Guha R, Petrus MN, et al. Selective targeting of JAK/STAT signaling is potentiated by Bcl-xL blockade in IL-2-dependent adult T-cell leukemia. Proc Natl Acad Sci U S A. 2015;112(40):12480–5.
Ishikawa C, Senba M, Mori N. Anti-adult T cell leukemia/lymphoma activity of cerdulatinib, a dual SYK/JAK kinase inhibitor. Int J Oncol. 2018;53(4):1681–90.
Yang H, Ma P, Cao Y, Zhang M, Li L, Wei J, et al. ECPIRM, a potential therapeutic agent for cutaneous T-cell lymphoma, inhibits cell proliferation and promotes apoptosis via a JAK/STAT pathway. Anti Cancer Agents Med Chem. 2018;18(3):401–11.
•• Moskowitz AJ, Jacobsen E, Ruan J, Schatz JH, Obadi O,Motylinski K, et al. Durable responses observed with JAK inhibition in T-cell lymphomas. ASH annual meeting; 2018 Dec 1–4; San Diego. [Abstract 2922]. Clinical evidence of the efficacy of JAK inhibition in TCL.
Waldmann TA, Chen J. Disorders of the JAK/STAT pathway in T cell lymphoma pathogenesis: implications for immunotherapy. Annu Rev Immunol. 2017;35:533–50.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on T-Cell and Other Lymphoproliferative Malignancies
Rights and permissions
About this article
Cite this article
Shouse, G., Nikolaenko, L. Targeting the JAK/STAT Pathway in T Cell Lymphoproliferative Disorders. Curr Hematol Malig Rep 14, 570–576 (2019). https://doi.org/10.1007/s11899-019-00545-5
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11899-019-00545-5