Abstract
Type 1 diabetes is an autoimmune disease with a complex polygenic inheritance. Until recently, only three susceptibility genes had been reproducibly identified, namely HLA, INSVNTR, and CTLA4. During the past 7 years, a number of new putative susceptibility genes have been isolated from both human and animal models of the disease. We present eight genes implicated in type 1 diabetes etiology and discuss them in relation to the pathogenesis of the disease: VDR, IL6, IL12B, AIRE, FOXP3, B2m, Cblb, and Lyp/Ian4l1.
Similar content being viewed by others
References and Recommended Reading
Pociot F, McDermott MF: Genetics of type 1 diabetes mellitus Genes Immun 2002, 3:235–249.
McDermott MF, Ramachandran A, Ogunkolade BW, et al.: Allelic variation in the vitamin D receptor influences susceptibility to IDDM in Indian Asians. Diabetologia 1997, 40:971–975.
Pani MA, Knapp M, Donner H, et al.: Vitamin D receptor allele combinations influence genetic susceptibility to type 1 diabetes in Germans. Diabetes 2000, 49:504–507.
Motohashi Y, Yamada S, Yanagawa T, et al.: Vitamin D receptor gene polymorphism affects onset pattern of type 1 diabetes. J Clin Endocrinol Metab 2003 88:3137–3140.
Hayes CE, Nashold FE, Spach KM, Pedersen LB: The immunological functions of the vitamin D endocrine system. Cell Mol Biol (Noisy-le-grand) 2003, 49:277–300.
Hitman GA, Mannan N, McDermott MF, et al.: Vitamin D receptor gene polymorphisms influence insulin secretion in Bangladeshi Asians. Diabetes 1998, 47:688–690.
Norman AW, Frankel JB, Heldt AM, Grodsky GM: Vitamin D deficiency inhibits pancreatic secretion of insulin. Science 1980, 209:823–825.
Kristiansen OP, Nolsoe RL, Larsen L, et al.: Association of a functional 17beta-estradiol sensitive IL6-174G/C promoter polymorphism with early-onset type 1 diabetes in females. Hum Mol Genet 2003, 12:1101–1110. This study demonstrates significant LD of IL6 to T1D in a Danish cohort and demonstrates that the entire LD is exclusively found among female persons. The finding is supported by functional data.
Jahromi MM, Millward BA, Demaine AG: A polymorphism in the promoter region of the gene for interleukin. J Interferon Cytokine Res 2000, 20:885–888.
Costanzo C, Piacentini G, Vicentini L, et al.: Cell-specific differences in the regulation of IL-6 expression by PMA. Biochem Biophys Res Commun 1999, 260:577–581.
Terry CF, Loukaci V, Green FR: Cooperative influence of genetic polymorphisms on interleukin 6 transcriptional regulation. J Biol Chem 2000, 275:18138–18144.
Dawson E, Abecasis GR, Bumpstead S, et al.: A first-generation linkage disequilibrium map of human chromosome 22. Nature 2002, 418:544–548.
Ishihara K, Hirano T: IL-6 in autoimmune disease and chronic inflammatory proliferative disease. Cytokine Growth Factor Rev 2002, 13:357–368.
Pasare C, Medzhitov R: Toll pathway-dependent blockade of CD4+CD25+ T cell-mediated suppression by dendritic cells. Science 2003, 299:1033–1036.
Morahan G, Huang D, Ymer SI, et al.: Linkage disequilibrium of a type 1 diabetes susceptibility locus with a regulatory IL12B allele. Nat Genet 2001, 27:218–221. Genetic linkage and LD to T1D of the IL12B locus is demonstrated. LD is confirmed on a new family material. The findings are supported by functional data.
Davoodi-Semiromi A, Yang JJ, She JX: IL-12p40 is associated with type 1 diabetes in Caucasian-American families. Diabetes 2002, 51:2334–2336.
Bergholdt R, Ghandil P, Johannesen J, et al.: Genetic and functional evaluation of an interleukin-12 polymorphism (IDDM18) in type 1 diabetes families. J Med Genet 2004, in press. http://jmg.bmjjournals.com/future/?ck=nck
Dahlman I, Eaves IA, Kosoy R, et al.: Parameters for reliable results in genetic association studies in common disease. Nat Genet 2002, 30:149–150. A study that failed to confirm the findings of Morahan et al. [15]. This study and Bergholdt et al. [17] led to the conclusion that IL12B, at the most, is a locus of minor importance.
Huang D, Cancilla MR, Morahan G: Complete primary structure, chromosomal localisation, and definition of polymorphisms of the gene encoding the human interleukin-12 p40 subunit. Genes Immun 2000, 1:515–520.
Seegers D, Zwiers A, Strober W, et al.: A TaqI polymorphism in the 3’UTR of the IL-12 p40 gene correlates with increased IL-12 secretion. Genes Immun 2002, 3:419–423.
Szelachowska M, Kretowski A, Kinalska I: Increased in vitro interleukin-12 production by peripheral blood in high-risk IDDM first degree relatives. Horm Metab Res 1997, 29:168–171.
Simpson PB, Mistry MS, Maki RA, et al.: Cuttine edge: diabetes-associated quantitative trait locus, Idd4, is responsible for the IL-12p40 overexpression defect in nonobese diabetic (NOD) mice. J Immunol 2003, 171:3333–3337.
Adorini L: Interleukin 12 and autoimmune diabetes. Nat Genet 2001, 27:131–132.
van Veen T, Crusius JB, Schrijver HM, et al.: Interleukin-12p40 genotype plays a role in the susceptibility to multiple sclerosis. Ann Neurol 2001, 50:275.
Florez JC, Hirschhorn J, Altshuler D: The inherited basis of diabetes mellitus: implications for the genetic analysis of complex traits. Annu Rev Genomics Hum Genet 2003, 4:257–291.
Nagamine K, Peterson P, Scott HS, et al.: Positional cloning of the APECED gene. Nat Genet 1997 17:393–398.
An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. The Finnish-German APECED Consortium. Autoimmune Polyendocrinopathy-Candidiasis-Ectodermal Dystrophy [no authors listed]. Nat Genet 1997, 17:399–403.
Pitkanen J, Peterson P: Autoimmune regulator: from loss of function to autoimmunity. Genes Immun 2003, 4:12–21.
Liston A, Lesage S, Wilson J, et al.: Aire regulates negative selection of organ-specific T cells. Nat Immunol 2003, 4:350–354. Aire deficiency is demonstrated to cause failure to delete the organspecific cells in the thymus
Ramsey C, Winqvist O, Puhakka L, et al.: Aire deficient mice develop multiple features of APECED phenotype and show altered immune response. Hum Mol Genet 2002, 11:397–409.
Anderson MS, Venanzi ES, Klein L, et al.: Projection of an immunological self shadow within the thymus by the aire protein. Science 2002, 298:1395–1401.
Nithiyananthan R, Heward JM, Allahabadia A, et al.: A heterozygous deletion of the autoimmune regulator (AIRE1) gene, autoimmune thyroid disease, and type 1 diabetes: no evidence for association. J Clin Endocrinol Metab 2000, 85:1320–1322. None of 235 patients with T1D were found to carry the mutation in exon 8 of the AIRE gene that accounts for more than 70% of mutant alleles in UK subjects with APECED.
Fontenot JD, Gavin MA, Rudensky AY: Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 2003, 4:330–336.
Khattri R, Cox T, Yasayko SA, Ramsdell F: An essential role for Scurfin in CD4+CD25+ T regulatory cells. Nat Immunol 2003, 4:337–342.
Hori S, Nomura T, Sakaguchi S: Control of regulatory T cell development by the transcription factor Foxp3. Science 2003, 299:1057–1061.
Zahorsky-Reeves JL, Wilkinson JE: A transgenic mouse strain with antigen-specific T cells (RAG1KO/sf/OVA) demonstrates that the scurfy (sf) mutation causes a defect in T-cell tolerization. Comp Med 2002, 52:58–62.
Chatila TA, Blaeser F, Ho N, et al.: JM2, encoding a fork head-related protein, is mutated in X-linked autoimmunity-allergic disregulation syndrome. J Clin Invest 2000, 106:R75-R81.
Schubert LA, Jeffery E, Zhang Y, et al.: Scurfin (FOXP3) acts as a repressor of transcription and regulates T cell activation. J Biol Chem 2001, 276:37672–37679.
Kasprowicz DJ, Smallwood PS, Tyznik AJ, Ziegler SF: Scurfin (FoxP3) controls T-dependent immune responses in vivo through regulation of CD4(+) T cell effector function. J Immunol 2003, 171:1216–1223.
Cucca F, Goy JV, Kawaguchi Y, et al.: A male-female bias in type 1 diabetes and linkage to chromosome Xp in MHC HLADR3-positive patients. Nat Genet 1998, 19:301–302.
Bassuny WM, Ihara K, Sasaki Y, et al.: A functional polymorphism in the promoter/enhancer region of the FOXP3/Scurfin gene associated with type 1 diabetes. Immunogenetics 2003, 55:149–156. This study showed significant association between the FOXP3/Scurfin gene and T1D in the Japanese population.
Ramsdell F: Foxp3 and natural regulatory T cells. Key to a cell lineage? Immunity 2003, 19:165–168. This is an excellent review on the role of FOXP3 in regulatory T cells.
Serreze DV, Bridgett M, Chapman HD, et al.: Subcongenic analysis of the Idd13 locus in NOD/Lt mice: evidence for several susceptibility genes including a possible diabetogenic role for beta 2-microglobulin. J Immunol 1998, 160:1472–1478.
Fox CJ, Paterson AD, Mortin-Toth SM, Danska JS: Two genetic loci regulate T cell-dependent islet inflammation and drive autoimmune diabetes pathogenesis. Am J Hum Genet 2000, 67:67–81. An early stage in diabetes pathogenesis in NOD mice, which behaves as a highly penetrant trait, is the T lymphocyte-dependent progression from a benign to a destructive stage of insulitis. This report demonstrates that both the Idd5 and Idd13 loci regulate this step.
Hamilton-Williams EE, Serreze DV, Charlton B, et al.: Transgenic rescue implicates beta2-microglobulin as a diabetes susceptibility gene in nonobese diabetic (NOD) mice. Proc Natl Acad Sci U S A 2001, 98:11533–11538. Using allelic reconstitution by transgenic rescue, this paper showed that NOD mice expressing the normal NOD B2m allele developed diabetes, whereas NOD mice expressing another murine or human allele were protected.
Yang Y, Bao M, Yoon JW: Intrinsic defects in the T-cell lineage results in natural killer T-cell deficiency and the development of diabetes in the nonobese diabetic mouse. Diabetes 2001, 50:2691–2699.
Beaudoin L, Laloux V, Novak J, et al.: NKT cells inhibit the onset of diabetes by impairing the development of pathogenic T cells specific for pancreatic beta cells. Immunity 2002, 17:725–736.
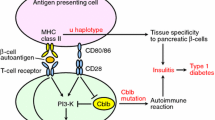
Yokoi N, Komeda K, Wang HY, et al.: Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet 2002, 31:391–394. Rat Iddm/kdp1 is identified as Cblb through positional cloning. The finding is confirmed by transgenic rescue and comparison to KO mouse strains.
Liu YC, Gu H: Cbl and Cbl-b in T-cell regulation. Trends Immunol 2002, 23:140–143.
Chiang YJ, Kole HK, Brown K, et al.: Cbl-b regulates the CD28 dependence of T-cell activation. Nature 2000, 403:216–220.
Bachmaier K, Krawczyk C, Kozieradzki I, et al.: Negative regulation of lymphocyte activation and autoimmunity by the molecular adaptor Cbl-b. Nature 2000, 403:211–216.
Zhang J, Bardos T, Li D, et al.: Cutting edge: regulation of T cell activation threshold by CD28 costimulation through targeting Cbl-b for ubiquitination. J Immunol 2002, 169:2236–2240.
Zhang W, Shao Y, Fang D, et al.: Negative regulation of T cell antigen receptor-mediated Crk-L-C3G signaling and cell adhesion by Cbl-b. J Biol Chem 2003, 278:23978–23983.
Ramanathan S, Poussier P: BB rat lyp mutation and type 1 diabetes. Immunol Rev 2001, 184:161–171.
Greiner DL, Handler ES, Nakano K, et al.: Absence of the RT-6 T cell subset in diabetes-prone BB/W rats. J Immunol 1986, 136:148–151.
Greiner DL, Mordes JP, Handler ES, et al.: Depletion of RT6.1+ T lymphocytes induces diabetes in resistant biobreeding/ Worcester (BB/W) rats. J Exp Med 1987, 166:461–475.
Burstein D, Mordes J, Greiner D, et al.: Prevention of diabetes in BB/Wor rat by single transfusion of spleen cells. Parameters that affect degree of protection. Diabetes 1989, 38:24–30.
Markholst H, Eastman S, Wilson D, et al.: Diabetes segregates as a single locus in crosses between inbred BB rats prone or resistant to diabetes. J Exp Med 1991, 174:297–300.
Awata T, Guberski DL, Like AA: Genetics of the BB rat: association of autoimmune disorders (diabetes, insulitis, and thyroiditis) with lymphopenia and major histocompatibility complex class II. Endocrinology 1995, 136:5731–5735.
Ramanathan S, Norwich K, Poussier P: Antigen activation rescues recent thymic emigrants from programmed cell death in the BB rat. J Immunol 1998, 160:5757–5764.
Pandarpurkar M, Wilson-Fritch L, Corvera S, et al.: Ian4 is required for mitochondrial integrity and T cell survival. Proc Natl Acad Sci U S A 2003, 100:10382–10387.
Hornum L, Rømer J, Markholst H: The diabetes-prone BB rat carries a frameshift mutation in Ian4, a positional candidate of Iddm1. Diabetes 2002, 51:1972–1979. Identified rat Iddm1/Lyp as Ian411 through a positional cloning approach. This was confirmed by MacMurray et al. [63].
MacMurray AJ, Moralejo DH, Kwitek AE, et al.: Lymphopenia in the BB rat model of type 1 diabetes is due to a mutation in a novel immune-associated nucleotide (Ian)-related gene. Genome Res 2002, 12:1029–1039.
Sandal T, Aumo L, Hedin L, et al.: Irod/Ian5: An inhibitor of gamma-radiation-and okadaic acid-induced apoptosis. Mol Biol Cell 2003, 14:3292–3304.
Hornum L, Kristensen OP, Pociot F, et al.: No association to type 1 diabetes of the human homologue of rat lyp (Iddm1) in the Danish population Diabetologia 2000, 43:A8.
Ueda H, Howson JM, Esposito L, et al.: Association of the T-cell regulatory gene CTLA4 with susceptibility to autoimmune disease. Nature 2003, 423:506–511.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hornum, L., Markholst, H. New autoimmune genes and the pathogenesis of type 1 diabetes. Curr Diab Rep 4, 135–142 (2004). https://doi.org/10.1007/s11892-004-0069-6
Issue Date:
DOI: https://doi.org/10.1007/s11892-004-0069-6