Abstract
Purpose of Review
This review summarizes the role of the microbiome in colorectal cancer (CRC) in the setting of immunotherapy and emphasizes the potential of microbiota-influencing strategies with a focus on the use of fecal microbiota transplant (FMT).
Recent Findings
Observations from preclinical and clinical studies suggest that the human gut microbiome is implicated in the CRC carcinogenesis and is integral in determining the clinical response and toxicity to immunotherapy. Among the therapeutic methods devised to exploit the microbiome, FMT is the most direct method and is backed by the highest level of evidence of efficacy in nonneoplastic disease settings. Furthermore, a favorable microbiome has the potential to overcome immunotherapy resistance and ameliorate immune-related adverse events (irAEs). To this end, clinical trials are underway to evaluate the potential of FMT and microbiota-augmented methods in the setting of immunotherapy in CRC.
Summary
Evidence from animal studies, retrospective studies, and smaller-scale prospective human studies have led to initiation of a number of microbiota-augmented clinical trials in CRC. Given the intimate relationship between the gut microbiota and the immune system as well as antitumor immune responses, potentiating immunotherapy and managing its toxicity are major areas of research in microbiota-augmented therapies in cancer. Therefore, evaluation of the patient microbiome as a routine part of clinical outcome analysis is warranted in future clinical trials.
Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. https://doi.org/10.3322/caac.21492.
Edwards BK, Ward E, Kohler BA, Eheman C, Zauber AG, Anderson RN, et al. Annual report to the nation on the status of cancer, 1975–2006, featuring colorectal cancer trends and impact of interventions (risk factors, screening, and treatment) to reduce future rates. Cancer. 2010;116(3):544–73. https://doi.org/10.1002/cncr.24760.
Garborg K, Holme Ø, Løberg M, Kalager M, Adami HO, Bretthauer M. Current status of screening for colorectal cancer. Ann Oncol. 2013;24(8):1963–72. https://doi.org/10.1093/annonc/mdt157.
Sargent D, Sobrero A, Grothey A, O'Connell MJ, Buyse M, Andre T, et al. Evidence for cure by adjuvant therapy in colon cancer: observations based on individual patient data from 20,898 patients on 18 randomized trials. J Clin Oncol. 2009;27(6):872–7. https://doi.org/10.1200/JCO.2008.19.5362.
Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17. https://doi.org/10.3322/caac.21220.
Diaz LA, Uram JN, Wang H, Bartlett B, Kemberling H, Eyring A, et al. Programmed death-1 blockade in mismatch repair deficient cancer independent of tumor histology. J Clin Oncol. 2016;34(15_suppl):3003. https://doi.org/10.1200/JCO.2016.34.15_suppl.3003.
Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–20. https://doi.org/10.1056/NEJMoa1500596.
Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz H-J, Morse MA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–91. https://doi.org/10.1016/S1470-2045(17)30422-9.
Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–57.
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–4. https://doi.org/10.1126/science.1129139.
Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–13. https://doi.org/10.1126/science.aan6733.
Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–18. https://doi.org/10.1200/JCO.2005.01.086.
Venderbosch S, Nagtegaal ID, Maughan TS, Smith CG, Cheadle JP, Fisher D, et al. Mismatch repair status and BRAF mutation status in metastatic colorectal cancer patients: a pooled analysis of the CAIRO, CAIRO2, COIN, and FOCUS studies. Clin Cancer Res. 2014;20(20):5322–30. https://doi.org/10.1158/1078-0432.CCR-14-0332.
Brahmer JR, Lacchetti C, Schneider BJ, Atkins MB, Brassil KJ, Caterino JM, et al. Management of immune-related adverse events in patients treated with immune checkpoint inhibitor therapy: American Society of Clinical Oncology clinical practice guideline. J Clin Oncol. 2018;36(17):1714–68. https://doi.org/10.1200/JCO.2017.77.6385.
Overman MJ, Lonardi S, Wong KYM, Lenz H-J, Gelsomino F, Aglietta M, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–9. https://doi.org/10.1200/JCO.2017.76.9901.
Kim D, Zeng MY, Núñez G. The interplay between host immune cells and gut microbiota in chronic inflammatory diseases. Exp Mol Med. 2017;49(5):e339-e. https://doi.org/10.1038/emm.2017.24.
Czesnikiewicz-Guzik M, Müller DN. Scientists on the spot: salt, the microbiome, and cardiovascular diseases. Cardiovasc Res. 2018;114(10):e72–e3. https://doi.org/10.1093/cvr/cvy171.
Jangi S, Gandhi R, Cox LM, Li N, von Glehn F, Yan R, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. https://doi.org/10.1038/ncomms12015.
Khanna S, Montassier E, Schmidt B, Patel R, Knights D, Pardi DS, et al. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment Pharmacol Ther. 2016;44(7):715–27. https://doi.org/10.1111/apt.13750.
Hansen JJ, Sartor RB. Therapeutic manipulation of the microbiome in IBD: current results and future approaches. Curr Treat Options Gastroenterol. 2015;13(1):105–20. https://doi.org/10.1007/s11938-014-0042-7.
Tierney BT, He Y, Church GM, Segal E, Kostic AD, Patel CJ. The predictive power of the microbiome exceeds that of genome-wide association studies in the discrimination of complex human disease. bioRxiv. 2020:2019.12.31.891978. https://doi.org/10.1101/2019.12.31.891978.
Salosensaari A, Laitinen V, Havulinna AS, Meric G, Cheng S, Perola M, et al. Taxonomic signatures of long-term mortality risk in human gut microbiota. medRxiv. 2020:2019.12.30.19015842. https://doi.org/10.1101/2019.12.30.19015842.
Garrett WS. Cancer and the microbiota. Science. 2015;348(6230):80–6. https://doi.org/10.1126/science.aaa4972.
Tsilimigras MCB, Fodor A, Jobin C. Carcinogenesis and therapeutics: the microbiota perspective. Nat Microbiol. 2017;2:17008. https://doi.org/10.1038/nmicrobiol.2017.8.
Di Domenico EG, Cavallo I, Pontone M, Toma L, Ensoli F. Biofilm producing Salmonella typhi: chronic colonization and development of gallbladder cancer. Int J Mol Sci. 2017;18(9):1887. https://doi.org/10.3390/ijms18091887.
Huang Y, Fan XG, Wang ZM, Zhou JH, Tian XF, Li N. Identification of helicobacter species in human liver samples from patients with primary hepatocellular carcinoma. J Clin Pathol. 2004;57(12):1273–7. https://doi.org/10.1136/jcp.2004.018556.
Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345(2):196–202. https://doi.org/10.1016/j.canlet.2013.08.016.
Ishaq S, Nunn L. Helicobacter pylori and gastric cancer: a state of the art review. Gastroenterol Hepatol Bed Bench. 2015;8(Suppl 1):S6–S14.
Gold JS, Bayar S, Salem RR. Association of Streptococcus bovis bacteremia with colonic neoplasia and extracolonic malignancy. Arch Surg. 2004;139(7):760–5. https://doi.org/10.1001/archsurg.139.7.760.
Mima K, Nakagawa S, Sawayama H, Ishimoto T, Imai K, Iwatsuki M, et al. The microbiome and hepatobiliary-pancreatic cancers. Cancer Lett. 2017;402:9–15. https://doi.org/10.1016/j.canlet.2017.05.001.
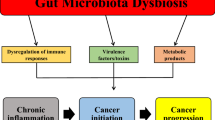
García-Castillo V, Sanhueza E, McNerney E, Onate SA, García A. Microbiota dysbiosis: a new piece in the understanding of the carcinogenesis puzzle. J Med Microbiol. 2016;65(12):1347–62. https://doi.org/10.1099/jmm.0.000371.
Riquelme E, Zhang Y, Zhang L, Montiel M, Zoltan M, Dong W, et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell. 2019;178(4):795–806.e12. https://doi.org/10.1016/j.cell.2019.07.008.
Flemer B, Lynch DB, Brown JMR, Jeffery IB, Ryan FJ, Claesson MJ, et al. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut. 2017;66(4):633–43. https://doi.org/10.1136/gutjnl-2015-309595.
Scott AJ, Alexander JL, Merrifield CA, Cunningham D, Jobin C, Brown R, et al. International cancer microbiome consortium consensus statement on the role of the human microbiome in carcinogenesis. Gut. 2019;68(9):1624–32. https://doi.org/10.1136/gutjnl-2019-318556.
Purcell RV, Pearson J, Aitchison A, Dixon L, Frizelle FA, Keenan JI. Colonization with enterotoxigenic Bacteroides fragilis is associated with early-stage colorectal neoplasia. PLoS One. 2017;12(2):e0171602-e. https://doi.org/10.1371/journal.pone.0171602.
Wu S, Rhee K-J, Albesiano E, Rabizadeh S, Wu X, Yen H-R, et al. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat Med. 2009;15(9):1016–22. https://doi.org/10.1038/nm.2015.
Boleij A, Hechenbleikner EM, Goodwin AC, Badani R, Stein EM, Lazarev MG, et al. The Bacteroides fragilis toxin gene is prevalent in the colon mucosa of colorectal cancer patients. Clin Infect Dis. 2015;60(2):208–15. https://doi.org/10.1093/cid/ciu787.
He Z, Gharaibeh RZ, Newsome RC, Pope JL, Dougherty MW, Tomkovich S, et al. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68(2):289–300. https://doi.org/10.1136/gutjnl-2018-317200.
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA, Michaud M, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–15. https://doi.org/10.1016/j.chom.2013.07.007.
Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42(2):344–55. https://doi.org/10.1016/j.immuni.2015.01.010.
Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S, et al. Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature. 2013;499(7456):97–101. https://doi.org/10.1038/nature12347.
Ma C, Han M, Heinrich B, Fu Q, Zhang Q, Sandhu M, et al. Gut microbiome-mediated bile acid metabolism regulates liver cancer via NKT cells. Science. 2018;360(6391):eaan5931. https://doi.org/10.1126/science.aan5931.
Kwa M, Plottel CS, Blaser MJ, Adams S. The intestinal microbiome and estrogen receptor-positive female breast cancer. J Natl Cancer Inst. 2016;108(8):djw029. https://doi.org/10.1093/jnci/djw029.
Pushalkar S, Hundeyin M, Daley D, Zambirinis CP, Kurz E, Mishra A, et al. The pancreatic cancer microbiome promotes oncogenesis by induction of innate and adaptive immune suppression. Cancer Discov. 2018;8(4):403–16. https://doi.org/10.1158/2159-8290.CD-17-1134.
Cronin M, Morrissey D, Rajendran S, El Mashad SM, van Sinderen D, O'Sullivan GC, et al. Orally administered bifidobacteria as vehicles for delivery of agents to systemic tumors. Mol Ther. 2010;18(7):1397–407. https://doi.org/10.1038/mt.2010.59.
Morrissey D, O'Sullivan GC, Tangney M. Tumour targeting with systemically administered bacteria. Curr Gene Ther. 2010;10(1):3–14. https://doi.org/10.2174/156652310790945575.
Xavier JB, Young VB, Skufca J, Ginty F, Testerman T, Pearson AT, et al. The cancer microbiome: distinguishing direct and indirect effects requires a systemic view. Trends Cancer. 2020;6(3):192–204. https://doi.org/10.1016/j.trecan.2020.01.004.
•• Yu T, Guo F, Yu Y, Sun T, Ma D, Han J, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170(3):548–63.e16. https://doi.org/10.1016/j.cell.2017.07.008This study demonstrates there is a dose-dependent relationship between F. nucleatum spp. abundance and progression from less to more malignant colorectal adenoma/adenocarcinomas.
•• Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med. 2019;25(6):968–76. https://doi.org/10.1038/s41591-019-0458-7This meta-analysis identified a microbiota signature from a set of geographically and technically heterogeneous studies, which was validated in each individual component study for diagnostic accuracy.
Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med. 2019;25(4):679–89. https://doi.org/10.1038/s41591-019-0406-6.
Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun. 2015;6:6528. https://doi.org/10.1038/ncomms7528.
Yu J, Feng Q, Wong SH, Zhang D, Liang QY, Qin Y, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–8. https://doi.org/10.1136/gutjnl-2015-309800.
Baxter NT, Ruffin MT, Rogers MAM, Schloss PD. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016;8(1):37. https://doi.org/10.1186/s13073-016-0290-3.
Thomas AM, Manghi P, Asnicar F, Pasolli E, Armanini F, Zolfo M, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med. 2019;25(4):667–78. https://doi.org/10.1038/s41591-019-0405-7.
Nakatsu G, Zhou H, Wu WKK, Wong SH, Coker OO, Dai Z, et al. Alterations in enteric virome are associated with colorectal cancer and survival outcomes. Gastroenterology. 2018;155(2):529–41.e5. https://doi.org/10.1053/j.gastro.2018.04.018.
Eklöf V, Löfgren-Burström A, Zingmark C, Edin S, Larsson P, Karling P, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. Int J Cancer. 2017;141(12):2528–36. https://doi.org/10.1002/ijc.31011.
García-González AP, Ritter AD, Shrestha S, Andersen EC, Yilmaz LS, Walhout AJM. Bacterial metabolism affects the C. elegans response to cancer chemotherapeutics. Cell. 2017;169(3):431–41.e8. https://doi.org/10.1016/j.cell.2017.03.046.
Viaud S, Saccheri F, Mignot G, Yamazaki T, Daillère R, Hannani D, et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science. 2013;342(6161):971–6. https://doi.org/10.1126/science.1240537.
Daillère R, Vétizou M, Waldschmitt N, Yamazaki T, Isnard C, Poirier-Colame V, et al. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity. 2016;45(4):931–43. https://doi.org/10.1016/j.immuni.2016.09.009.
Vande Voorde J, Sabuncuoğlu S, Noppen S, Hofer A, Ranjbarian F, Fieuws S, et al. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J Biol Chem. 2014;289(19):13054–65. https://doi.org/10.1074/jbc.M114.558924.
Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342(6161):967–70. https://doi.org/10.1126/science.1240527.
Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330(6005):831–5. https://doi.org/10.1126/science.1191175.
Nam Y-D, Kim HJ, Seo J-G, Kang SW, Bae J-W. Impact of pelvic radiotherapy on gut microbiota of gynecological cancer patients revealed by massive pyrosequencing. PLoS One. 2013;8(12):e82659-e. https://doi.org/10.1371/journal.pone.0082659.
Kim YS, Kim J, Park S-J. High-throughput 16S rRNA gene sequencing reveals alterations of mouse intestinal microbiota after radiotherapy. Anaerobe. 2015;33:1–7. https://doi.org/10.1016/j.anaerobe.2015.01.004.
Gerassy-Vainberg S, Blatt A, Danin-Poleg Y, Gershovich K, Sabo E, Nevelsky A, et al. Radiation induces proinflammatory dysbiosis: transmission of inflammatory susceptibility by host cytokine induction. Gut. 2018;67(1):97–107. https://doi.org/10.1136/gutjnl-2017-313789.
Sokol H, Adolph TE. The microbiota: an underestimated actor in radiation-induced lesions? Gut. 2018;67(1):1–2. https://doi.org/10.1136/gutjnl-2017-314279.
Panda A, Mehnert JM, Hirshfield KM, Riedlinger G, Damare S, Saunders T, et al. Immune activation and benefit from Avelumab in EBV-positive gastric cancer. J Natl Cancer Inst. 2018;110(3):316–20. https://doi.org/10.1093/jnci/djx213.
Host KM, Jacobs SR, West JA, Zhang Z, Costantini LM, Stopford CM, et al. Kaposi's sarcoma-associated herpesvirus increases PD-L1 and proinflammatory cytokine expression in human monocytes. mBio. 2017;8(5):e00917. https://doi.org/10.1128/mBio.00917-17.
Smola S. Immunopathogenesis of HPV-associated cancers and prospects for immunotherapy. Viruses. 2017;9(9):254. https://doi.org/10.3390/v9090254.
• Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359(6371):91–7. https://doi.org/10.1126/science.aan3706This study demonstrates the association between a specific bacterial species and immunotherapy response via an experiment involving FMT in gnotobiotic mice.
Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre M-L, et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science. 2018;359(6371):104–8. https://doi.org/10.1126/science.aao3290.
Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science. 2018;359(6371):97–103. https://doi.org/10.1126/science.aan4236.
Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28(6):1368–79. https://doi.org/10.1093/annonc/mdx108.
•• Frankel AE, Coughlin LA, Kim J, Froehlich TW, Xie Y, Frenkel EP, et al. Metagenomic shotgun sequencing and unbiased metabolomic profiling identify specific human gut microbiota and metabolites associated with immune checkpoint therapy efficacy in melanoma patients. Neoplasia. 2017;19(10):848–55. https://doi.org/10.1016/j.neo.2017.08.004This study shows that a specific microbiota phylum was associated with protection against immune-related colitis in the setting of anti-CTLA-4 treatment in metastatic melanoma.
Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. https://doi.org/10.1038/ncomms10391.
Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350(6264):1079–84. https://doi.org/10.1126/science.aad1329.
Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science. 2015;350(6264):1084–9. https://doi.org/10.1126/science.aac4255.
Peled JU, Devlin SM, Staffas A, Lumish M, Khanin R, Littmann ER, et al. Intestinal microbiota and relapse after hematopoietic-cell transplantation. J Clin Oncol. 2017;35(15):1650–9. https://doi.org/10.1200/JCO.2016.70.3348.
•• Gharaibeh RZ, Jobin C. Microbiota and cancer immunotherapy: in search of microbial signals. Gut. 2018;68(3):385–8. https://doi.org/10.1136/gutjnl-2018-317220This study identifies a microbiota signature in a colon cancer animal model that is associated with favorable immunotherapy response.
Tanoue T, Morita S, Plichta DR, Skelly AN, Suda W, Sugiura Y, et al. A defined commensal consortium elicits CD8 T cells and anticancer immunity. Nature. 2019;565(7741):600–5. https://doi.org/10.1038/s41586-019-0878-z.
Gough E, Shaikh H, Manges AR. Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis. 2011;53(10):994–1002. https://doi.org/10.1093/cid/cir632.
Kao D, Roach B, Silva M, Beck P, Rioux K, Kaplan GG, et al. Effect of oral capsule- vs colonoscopy-delivered fecal microbiota transplantation on recurrent Clostridium difficile infection: a randomized clinical trial. JAMA. 2017;318(20):1985–93. https://doi.org/10.1001/jama.2017.17077.
Kelly CR, Ihunnah C, Fischer M, Khoruts A, Surawicz C, Afzali A, et al. Fecal microbiota transplant for treatment of Clostridium difficile infection in immunocompromised patients. Am J Gastroenterol. 2014;109(7):1065–71. https://doi.org/10.1038/ajg.2014.133.
Karakan T. Fecal microbiota transplant in immunocompromised patients: encouraging results in a vulnarable population. Turk J Gastroenterol. 2014;25(3):346. https://doi.org/10.5152/tjg.2014.0013.
D'Haens GR, Jobin C. Fecal microbial transplantation for diseases beyond recurrent clostridium difficile infection. Gastroenterology. 2019;157(3):624–36. https://doi.org/10.1053/j.gastro.2019.04.053.
McQuade JL, Daniel CR, Helmink BA, Wargo JA. Modulating the microbiome to improve therapeutic response in cancer. Lancet Oncol. 2019;20(2):e77–91. https://doi.org/10.1016/S1470-2045(18)30952-5.
• van Lier YF, Davids M, Haverkate NJE, de Groot PF, Donker ML, Nur E, et al. Fecal microbiota transplantation can cure steroid-refractory intestinal graft-versus-host disease. Biol Blood Marrow Transplant. 2019;25(3, Supplement):S241. https://doi.org/10.1016/j.bbmt.2018.12.237This prospective single-arm study demonstrates the efficacy of FMT in treating adverse events driven by an immunlogic process in the setting of hematologic cancer.
Qi X, Li X, Zhao Y, Wu X, Chen F, Ma X, et al. Treating steroid refractory intestinal acute graft-vs.-host disease with fecal microbiota transplantation: apilot study. Front Immunol. 2018;9:2195. https://doi.org/10.3389/fimmu.2018.02195.
•• Wang Y, Wiesnoski DH, Helmink BA, Gopalakrishnan V, Choi K, DuPont HL, et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor-associated colitis. Nat Med. 2018;24(12):1804–8. https://doi.org/10.1038/s41591-018-0238-9This is the first case series to demonstrate the efficacy of FMT in the treatment of irAE refractory to systemic immunosuppressive therapy.
Acknowledgements
The editors would like to thank Dr. Sakti Chakrabarti for taking the time to review this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Robin Park and Shahid Umar each declare no potential conflicts of interest.
Anup Kasi has received financial support, paid to his institution, from TESARO, Halozyme, Geistlich Pharma, Astellas Pharma, and Rafael Pharmaceuticals, and honoraria from OncLive.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Basic Science Foundations in Colorectal Cancer
Rights and permissions
About this article
Cite this article
Park, R., Umar, S. & Kasi, A. Immunotherapy in Colorectal Cancer: Potential of Fecal Transplant and Microbiota-Augmented Clinical Trials. Curr Colorectal Cancer Rep 16, 81–88 (2020). https://doi.org/10.1007/s11888-020-00456-1
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11888-020-00456-1