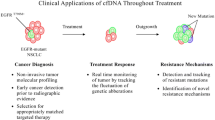
Opinion statement
Isolation and analysis of circulating tumor DNA (ctDNA) have emerged as an effective and promising tool for genomic profiling in non-small cell lung cancer. Analysis of ctDNA can be particularly useful in situations where tissue biopsy is not safely obtainable due to poor physical condition or inaccessible tumor biopsy location. In addition to identifying oncogenic driver mutations which can be treated with targetable therapy in the treatment naïve advanced non-small cell lung cancer (NSCLC) setting, ctDNA is being utilized in novel ways including monitoring during an advanced NSCLC patient’s treatment course (real-time monitoring), determining mechanisms of resistance and, lastly, as a tool to identify minimal residual disease in early-stage NSCLC. Recent research demonstrates that ctDNA testing can provide a useful adjunct to tissue genotyping in NSCLC. Utilization of ctDNA into routine clinical practice for NSCLC should be strongly considered.

Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Chan BA, Hughes BG. Targeted therapy for non-small cell lung cancer: current standards and the promise of the future. Transl Lung Cancer Res. 2015;4(1):36–54. https://doi.org/10.3978/j.issn.2218-6751.2014.05.01.
Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–39.
Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–500.
National Comprehensive Cancer Network. Non-small cell lung cancer (Version 2.2019). https://www.nccn.org/professionals/physician_gls/pdf/nscl.pdf. Accessed 9 Jan 2019.
•• Rolfo C, Mack PC, Scagliotti GV, Baas P, Barlesi F, Bivona TG, et al. Liquid biopsy for advanced non-small cell lung cancer (NSCLC): a statement paper from the IALSC. J Thorac Oncol. 2018;13(9):1248–68. https://doi.org/10.1016/j.jtho.2018.05.030 Evidence-based recommendations regarding the use of liquid biopsy in NSCLC provided by a multidisciplinary panel of experts in thoracic oncology.
Uozu S, Imaizumi K, Yamaguchi T, Goto Y, Kawada K, Minezawa T, et al. Feasibility of tissue re-biopsy in non-small cell lung cancers resistant to previous epidermal growth factor receptor tyrosine kinase inhibitor therapies. BMC Pulm Med. 2017;17(1):175. https://doi.org/10.1186/s12890-017-0514-3.
Sacher AG, Paweletz C, Dahlberg SE, Alden RS, O’Connell A, Feeney N, et al. Prospective validation of rapid plasma genotyping for the detection of EGFR and KRAS mutations in advanced lung cancer. JAMA Oncol. 2016;2(8):1014–22. https://doi.org/10.1001/jamaoncol.2016.0173.
Zhang J, Fujimoto J, Zhang J, Wedge DC, Song X, Zhang J, et al. Intratumor heterogeneity in localized lung adenocarcinomas delineated by multiregion sequencing. Science. 2014;346(6206):256–9. https://doi.org/10.1126/science.1256930.
Kim EY, Cho EN, Park HS, Kim A, Hong JY, Lim S, et al. Genetic heterogeneity of actionable genes between primary and metastatic tumor in lung adenocarcinoma. BMC Cancer. 2016;16:27. https://doi.org/10.1186/s12885-016-2049-z.
Han HS, Eom DW, Kim JH, Kim KH, Shin HM, An JY, et al. EGFR mutation status in primary lung adenocarcinomas and corresponding metastatic lesions: discordance in pleural metastases. Clin Lung Cancer. 2011;12(6):380–6. https://doi.org/10.1016/j.cllc.2011.02.006.
Sacher AG, Komatsubara KM, Oxnard GR. Application of plasma genotyping technologies in non-small cell lung cancer: a practical review. J Thorac Oncol. 2017;12(9):1344–56. https://doi.org/10.1016/j.jtho.2017.05.022.
Passiglia F, Rizzo S, Rolfo C, Galvano A, Bronte E, Incorvaia L, et al. Metastatic site location influences the diagnostic accuracy of ctDNA EGFR-mutation testing in NSCLC patients: a pooled analysis. Curr Cancer Drug Targets. 2018;18(7):697–705. https://doi.org/10.2174/1568009618666180308125110.
Boettcher S, Ebert BL. Clonal hematopoiesis of indeterminate potential. J Clin Oncol. 2018:JCO2018793588;37:419–22. https://doi.org/10.1200/JCO.2018.79.3588.
Hu Y, Ulrich BC, Supplee J, Kuang Y, Lizotte PH, Feeney NB, et al. False-positive plasma genotyping due to clonal hematopoiesis. Clin Cancer Res. 2018;24(18):4437–43. https://doi.org/10.1158/1078-0432.CCR-18-0143.
Slavin TP, Banks KC, Chudova D, Oxnard GR, Odegaard JI, Nagy RJ, et al. Identification of incidental germline mutations in patients with advanced solid tumors who underwent cell-free circulating tumor DNA sequencing. J Clin Oncol. 2018:JCO1800328;36:3459–65. https://doi.org/10.1200/JCO.18.00328.
Sorber L, Zwaenepoel K, De Winne K, Van Casteren K, Augustus E, Jacobs J, et al. A multicenter study to assess EGFR mutational status in plasma: focus on an optimized workflow for liquid biopsy in a clinical setting. Cancers (Basel). 2018;10(9):E290. https://doi.org/10.3390/cancers10090290.
Wu YL, Lee V, Liam CK, Lu S, Park K, Srimuninnimit V, et al. Clinical utility of a blood-based EGFR mutation test in patients receiving first-line erlotinib therapy in the ENSURE, FASTACT-2, and ASPIRATION studies. Lung Cancer. 2018;126:1–8. https://doi.org/10.1016/j.lungcan.2018.10.004.
Malapelle U, Sirera R, Jantus-Lewintre E, Reclusa P, Calabuig-Farinas S, Blasco A, et al. Profile of the Roche cobas® EGFR mutation test v2 for non-small cell lung cancer. Expert Rev Mol Diagn. 2017;17(3):209–15. https://doi.org/10.1080/14737159.2017.1288568.
Oxnard GR, Paweletz CP, Kuang Y, Mach SL, O’Connell A, Messineo MM, et al. Noninvasive detection of response and resistance in EGFR-mutated lung cancer using quantitative next-generation genotyping of cell-free plasma DNA. Clin Cancer Res. 2014;20(6):1698–705. https://doi.org/10.1158/1078-0432.CCR-13-2482.
Zhang R, Chen B, Tong X, Wang Y, Wang C, Jin J, et al. Diagnostic accuracy of droplet digital PCR for detection of EGFR T790M mutation in circulating tumor DNA. Cancer Manag Res. 2018;10:1209–18. https://doi.org/10.2147/CMAR.S161382.
Taniguchi K, Uchida J, Nishino K, Kumagai T, Okuyama T, Okami J, et al. Quantitative detection of EGFR mutations in circulating tumor DNA derived from lung adenocarcinomas. Clin Cancer Res. 2011;17(24):7808–15. https://doi.org/10.1158/1078-0432.CCR-11-1712.
Thress KS, Brant R, Carr TH, Dearden S, Jenkins S, Brown H, et al. EGFR mutation detection in ctDNA from NSCLC patient plasma: a cross-platform comparison of leading technologies to support the clinical development of AZD9291. Lung Cancer. 2015;90(3):509–15. https://doi.org/10.1016/j.lungcan.2015.10.004.
Zhou C, Wang M, Cheng Y, Chen Y, Ye X, Sun Y, et al. Detection of EGFR T790M in Asia-Pacific patients (pts) with EGFR mutation-positive advanced non-small cell lung cancer (NSCLC): circulating tumour (ct) DNA analysis across 3 platforms. Ann Oncol. 2017;28(suppl_5):v460–96.
Rozenblum AB, Ilouze M, Dudnik E, Dvir A, Soussan-Gutman L, Geva S, et al. Clinical impact of hybrid capture-based next-generation sequencing on changes in treatment decisions in lung cancer. J Thorac Oncol. 2017;12(2):258–68. https://doi.org/10.1016/j.jtho.2016.10.021.
Aggarwal C, Thompson JC, Black TA, Katz SI, Fan R, Yee SS, et al. Clinical implications of plasma-based genotyping with the delivery of personalized therapy in metastatic non-small cell lung cancer. JAMA Oncol. 2018. https://doi.org/10.1001/jamaoncol.2018.4305 Prospective cohort study of advanced NSCLC patients demonstrating that plasma-based NGS testing increased detection of targetable alterations and improved delivery of targeted therapy.
Douillard JY, Ostoros G, Cobo M, Ciuleanu T, Cole R, McWalter G, et al. Gefitinib treatment in EGFR mutated caucasian NSCLC: circulating-free tumor DNA as a surrogate for determination of EGFR status. J Thorac Oncol. 2014;9(9):1345–53. https://doi.org/10.1097/JTO.0000000000000263.
Wang Z, Cheng Y, An T, Gao H, Wang K, Zhou Q, et al. Detection of EGFR mutations in plasma circulating tumour DNA as a selection criterion for first-line gefitinib treatment in patients with advanced lung adenocarcinoma (BENEFIT): a phase 2, single-arm, multicentre clinical trial. Lancet Respir Med. 2018;6(9):681–90. https://doi.org/10.1016/S2213-2600(18)30264-9.
Gray J, Okamoto I, Sriuranpong V, Vansteenkiste J, Imamura F, Lee JS, et al. Osimertinib vs SoC EGFR-TKI as first-line treatment in patients with EGFRm advanced NSCLC (FLAURA): plasma ctDNA analysis. J Thorac Oncol. 2017;12(11):S1754–5. https://doi.org/10.1016/j.jtho.2017.09.348.
Zheng D, Ye X, Zhang MZ, Sun Y, Wang JY, Ni J, et al. Plasma EGFR T790M ctDNA status is associated with clinical outcome in advanced NSCLC patients with acquired EGFR-TKI resistance. Sci Rep. 2016;6:20913. https://doi.org/10.1038/srep20913.
Ramalingam SS, Cheng Y, Zhou C, Ohe Y, Imamura F, Cho BC, et al. Mechanisms of acquired resistance to first-line osimertinib: preliminary data from the phase III FLAURA study. Ann Oncol. 2018;29(suppl_9):ix173–8.
• Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(28):3375–82. https://doi.org/10.1200/JCO.2016.66.7162 Retrospective study of EGFR-mutated NSCLC patients with acquired EGFR-TKI resistance, demonstrating comparable ORR and PFS among those with plasma T790M-positive and tumor T790M-positive results.
Dagogo-Jack I, Brannon AR, Ferris LA, Campbell CD, Lin JJ, Schultz KR, et al. Tracking the evolution of resistance to ALK tyrosine kinase inhibitors through longitudinal analysis of circulating tumor DNA. JCO Precis Oncol. 2018;2018:1–14. https://doi.org/10.1200/PO.17.00160.
Shaw AT, Martini JF, Besse B, Bauer TM, Lin CC, Soo RA, et al. Efficacy of lorlatinib in patients (pts) with advanced ALK-positive non-small cell lung cancer (NSCLC) and ALK kinase domain mutations. Cancer Res. 2018;78(13 Suppl):Abstract nr CT044.
Burstein HJ, Krilov L, Aragon-Ching JB, Baxter NN, Chiorean EG, Chow WA, et al. Clinical cancer advances 2017: annual report on progress against cancer from the American Society of Clinical Oncology. J Clin Oncol. 2017;35(12):1341–67. https://doi.org/10.1200/JCO.2016.71.5292.
Lindeman NI, Cagle PT, Aisner DL, Arcila ME, Beasley MB, Bernicker EH, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: guideline from the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch Pathol Lab Med. 2018;142(3):321–46. https://doi.org/10.5858/arpa.2017-0388-CP.
Abbosh C, Birkbak NJ, Wilson GA, Jamal-Hanjani M, Constantin T, Salari R, et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature. 2017;545(7655):446–51. https://doi.org/10.1038/nature22364.
Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early detection of molecular residual disease in localized lung cancer by circulating tumor DNA profiling. Cancer Discov. 2017;7(12):1394–403. https://doi.org/10.1158/2159-8290.CD-17-0716.
Abbosh C, Birkbak NJ, Swanton C. Early stage NSCLC – challenges to implementing ctDNA-based screening and MRD detection. Nat Rev Clin Oncol. 2018;15(9):577–86. https://doi.org/10.1038/s41571-018-0058-3.
Gandara DR, Paul SM, Kowanetz M, Schleifman E, Zou W, Li Y, et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat Med. 2018;24(9):1441–8. https://doi.org/10.1038/s41591-018-0134-3.
Kim ES, Velcheti V, Mekhail T, Leal TA, Dowell JE, Tsai ML, et al. Primary efficacy results from B-F1RST, a prospective phase II trial evaluating blood-based tumour mutational burden (bTMB) as a predictive biomarker for atezolizumab (atezo) in 1L non-small cell lung cancer (NSCLC). Ann Oncol. 2018;29(suppl_5):Abstr LBA55.
Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018;24(24):6212–22. https://doi.org/10.1158/1078-0432.CCR-18-0386.
Khagi Y, Goodman AM, Daniels GA, Patel SP, Sacco AG, Randall JM, et al. Hypermutated circulating tumor DNA: correlation with response to checkpoint inhibitor-based immunotherapy. Clin Cancer Res. 2017;23(19):5729–36. https://doi.org/10.1158/1078-0432.CCR-17-1439.
Reckamp KL, Melnikova VO, Karlovich C, Sequist LV, Camidge DR, Wakelee H, et al. A highly sensitive and quantitative test platform for detection of NSCLC EGFR mutations in urine and plasma. J Thorac Oncol. 2016;11(10):1690–700. https://doi.org/10.1016/j.jtho.2016.05.035.
Husain H, Melnikova VO, Kosco K, Woodward B, More S, Pingle SC, et al. Monitoring daily dynamics of early tumor response to targeted therapy by detecting circulating tumor DNA in urine. Clin Cancer Res. 2017;23(16):4716–23. https://doi.org/10.1158/1078-0432.CCR-17-0454.
Jiang BY, Yangsi LI, Chuai S, Zhang Z, Yang JJ, Zhong W, et al. NGS to reveal heterogeneity between cerebrospinal fluid and plasma ctDNA among non-small cell lung cancer patients with leptomeningeal carcinomatosis. J Clin Oncol. 2017;35(15):suppl:9022–9022. https://doi.org/10.1200/JCO.2017.35.15_suppl.9022.
Acknowledgements
The authors thank Joseph Pinto, PhD, for his cooperation in the creation of the figure.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Katherine A. Scilla declares that she has no conflict of interest.
Christian Rolfo has received speaker’s honoraria from Guardant Health, MSD, and Novartis; has received non-financial support from OncoDNA (exosomes research collaboration) and Mylan (scientific advisor); and is currently Vice President of the International Society of Liquid Biopsy (ISLB).
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Lung Cancer
Rights and permissions
About this article
Cite this article
Scilla, K.A., Rolfo, C. The Role of Circulating Tumor DNA in Lung Cancer: Mutational Analysis, Diagnosis, and Surveillance Now and into the Future. Curr. Treat. Options in Oncol. 20, 61 (2019). https://doi.org/10.1007/s11864-019-0653-2
Published:
DOI: https://doi.org/10.1007/s11864-019-0653-2