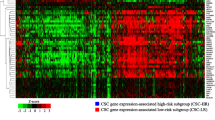
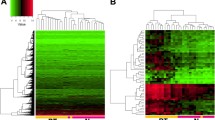
Opinion statement
Head and neck squamous cell carcinoma (HNSCC) is the sixth leading cancer by incidence worldwide and considering the recent EUROCARE-5 population-based study the 5-year survival rate of HNSCC patients in Europe ranges between 69% in localized cases and 34% in patients with regional involvement. The development of high-throughput gene expression assays in the last two decades has provided the invaluable opportunity to improve our knowledge on cancer biology and to identify predictive signatures in the most deeply analyzed malignancies, such as hematological and breast cancers. At variance, till 2010, the number of reliable reports referring gene expression data related to HSNCC biology and prediction was quite limited. A critical revision of the literature reporting gene expression data in HNSCC indicated that in the last 6 years, there were new important studies with a relevant increase in the sample size and a more accurate selection of cases, the publication of a growing number of studies applying a computational integration (meta-analysis) of different microarray datasets addressing similar clinical/biological questions, the increased use of molecular sub-classification of tumors according to their gene expression, and the release of the publicly available largest dataset in HNSCC by The Cancer Genome Atlas (TCGA) consortium. Overall, also for this disease, it become evident that the expression analysis of the entire transcriptome has been enabling to achieve the identification of promising molecular signatures for (i) disclosure of the biology behind carcinogenesis with special focus on the HPV-related one, (ii) prediction of tumor recurrence or metastasis development, (iii) identification of subgroups of tumors with different biology and associated prognosis, and (iv) prediction of outcome and/or response to therapy. The increasing awareness of the relevance of strict collaboration among clinicians and translational researchers would in a near future enable the application of a personalized HNSCCs patients’ treatment in the clinical practice based also on gene expression signatures.
Similar content being viewed by others
References and Recommended Reading
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Gatta G, Botta L, Sánchez MJ, Anderson LA, Pierannunzio D, Licitra L. EUROCARE Working Group. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: The EUROCARE-5 population-based study. Eur J Cancer. 2015.
Bose P, Brockton NT, Dort JC. Head and neck cancer: from anatomy to biology. Int J Cancer. 2013;133(9):2013–23.
Kang H, Kiess A, Chung CH. Emerging biomarkers in head and neck cancer in the era of genomics. Nat Rev Clin Oncol. 2015;12(1):11–26.
Macgregor PF, Squire JA. Application of microarrays to the analysis of gene expression in cancer. Clin Chem. 2002;48(8):1170–7.
Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS. Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res. 2011;13(3):212.
Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270(5235):467–70.
Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63.
Kumar S, Vo AD, Qin F, Li H. Comparative assessment of methods for the fusion transcripts detection from RNA-seq data. Sci Rep. 2016;6:21597.
Zhao S, Fung-Leung W-P, Bittner A, Ngo K, Liu X. Comparison of RNA-seq and microarray in transcriptome profiling of activated T cells. PLoS One. 2014;9(1):e78644.
Lallemant B, Evrard A, Chambon G, Sabra O, Kacha S, Lallemant JG, Lumbroso S, Brouillet JP. Gene expression profiling in head and neck squamous cell carcinoma: clinical perspectives. Head Neck. 2010;32(12):1712–9.
Ramasamy A, Mondry A, Holmes CC, Altman DG. Key issues in conducting a meta-analysis of gene expression microarray datasets. PLoS Med. 2008;5(9):e184.
•• Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–82. doi:10.1038/nature14129. This represents the larger study conducted in HNSCC in terms of number of cases and “omics” methods analyzed
Dobbin KK, Simon RM. Sample size planning for developing classifiers using high-dimensional DNA microarray data. Biostatistics. 2007;8(1):101–17.
Cromer A, Carles A, Millon R, Ganguli G, Chalmel F, Lemaire F, Young J, Dembélé D, Thibault C, Muller D, Poch O, Abecassis J, Wasylyk B. Identification of genes associated with tumorigenesis and metastatic potential of hypopharyngeal cancer by microarray analysis. Oncogene. 2004;23(14):2484–98.
Kuriakose MA, Chen WT, He ZM, Sikora AG, Zhang P, Zhang ZY, Qiu WL, Hsu DF, McMunn-Coffran C, Brown SM, Elango EM, Delacure MD, Chen FA. Selection and validation of differentially expressed genes in head and neck cancer. Cell Mol Life Sci. 2004;61(11):1372–83.
Dysvik B, Vasstrand EN, Løvlie R, Elgindi OA, Kross KW, Aarstad HJ, Johannessen AC, Jonassen I, Ibrahim SO. Gene expression profiles of head and neck carcinomas from Sudanese and Norwegian patients reveal common biological pathways regardless of race and lifestyle. Clin Cancer Res. 2006;12(4):1109–20.
Kondoh N, Ohkura S, Arai M, Hada A, Ishikawa T, Yamazaki Y, Shindoh M, Takahashi M, Kitagawa Y, Matsubara O, Yamamoto M. Gene expression signatures that can discriminate oral leukoplakia subtypes and squamous cell carcinoma. Oral Oncol. 2007;43(5):455–62.
Ye H, Yu T, Temam S, Ziober BL, Wang J, Schwartz JL, Mao L, Wong DT, Zhou X. Transcriptomic dissection of tongue squamous cell carcinoma. BMC Genomics. 2008;9:69.
Chen C, Méndez E, Houck J, Fan W, Lohavanichbutr P, Doody D, Yueh B, Futran ND, Upton M, Farwell DG, Schwartz SM, Zhao LP. Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomark Prev. 2008;17(8):2152–62.
Estilo CL, O-charoenrat P, Talbot S, Socci ND, Carlson DL, Ghossein R, Williams T, Yonekawa Y, Ramanathan Y, Boyle JO, Kraus DH, Patel S, Shaha AR, Wong RJ, Huryn JM, Shah JP, Singh B. Oral tongue cancer gene expression profiling: identification of novel potential prognosticators by oligonucleotide microarray analysis. BMC Cancer. 2009;9:11.
Saleh A, Zain RB, Hussaini H, Ng F, Tanavde V, Hamid S, Chow AT, Lim GS, Abraham MT, Teo SH, Cheong SC. Transcriptional profiling of oral squamous cell carcinoma using formalin-fixed paraffin-embedded samples. Oral Oncol. 2010;46(5):379–86.
Saintigny P, Zhang L, Fan YH, El-Naggar AK, Papadimitrakopoulou VA, Feng L, Lee JJ, Kim ES, Ki Hong W, Mao L. Gene expression profiling predicts the development of oral cancer. Cancer Prev Res (Phila). 2011;4(2):218–29.
Sun W, Gaykalova DA, Ochs MF, Mambo E, Arnaoutakis D, Liu Y, Loyo M, Agrawal N, Howard J, Li R, Ahn S, Fertig E, Sidransky D, Houghton J, Buddavarapu K, Sanford T, Choudhary A, Darden W, Adai A, Latham G, Bishop J, Sharma R, Westra WH, Hennessey P, Chung CH, Califano JA. Activation of the NOTCH pathway in head and neck cancer. Cancer Res. 2014;74(4):1091–104.
Masterson L, Sorgeloos F, Winder D, Lechner M, Marker A, Malhotra S, Sudhoff H, Jani P, Goon P, Sterling J. Deregulation of SYCP2 predicts early stage human papillomavirus-positive oropharyngeal carcinoma: a prospective whole transcriptome analysis. Cancer Sci. 2015;106(11):1568–75.
Yan L, Zhan C, Wu J, Wang S. Expression profile analysis of head and neck squamous cell carcinomas using data from the cancer genome atlas. Mol Med Rep. 2016;13(5):4259–65.
• Slebos RJ, Yi Y, Ely K, Carter J, Evjen A, Zhang X, Shyr Y, Murphy BM, Cmelak AJ, Burkey BB, Netterville JL, Levy S, Yarbrough WG, Chung CH. Gene expression differences associated with human papillomavirus status in head and neck squamous cell carcinoma. Clin Cancer Res. 2006;12(3):701–9. First signature able to stratify patients based on HPV status
Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, Kelsey KT, Turek LP, Ahlquist P. Fundamental differences in cell cycles deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancer. Cancer Res. 2007;67(10):4605–19.
Schlecht NF, Burk RD, Adrien L, Dunne A, Kawachi N, Sarta C, Chen Q, Brandwein-Gensler M, Prystowsky MB, Childs G, Smith RV, Belbin TJ. Gene expression profiles in HPV-infected head and neck cancer. J Pathol. 2007;213(3):283–93.
Lohavanichbutr P, Houck J, Fan W, Yueh B, Mendez E, Futran N, Doody DR, Upton MP, Farwell DG, Schwartz SM, Zhao LP, Chen C. Genomewide gene expression profiles of HPV-positive and HPV-negative oropharyngeal cancer: potential implications for treatment choices. Arch Otolaryngol Head Neck Surg. 2009;135(2):180–8.
Wichmann G, Rosolowski M, Krohn K, Kreuz M, Boehm A, Reiche A, Scharrer U, Halama D, Bertolini J, Bauer U, Holzinger D, Pawlita M, Hess J, Engel C, Hasenclever D, Scholz M, Ahnert P, Kirsten H, Hemprich A, Wittekind C, Herbarth O, Horn F, Dietz A, Loeffler M, Leipzig Head and Neck Group (LHNG). The role of HPV RNA transcription, immune response-related gene expression and disruptive TP53 mutations in diagnostic and prognostic profiling of head and neck cancer. Int J Cancer. 2015;137(12):2846–57.
Zhang Y, Koneva LA, Virani S, Arthur AE, Virani A, Hall PB, Warden CD, Carey TE, Chepeha DB, Prince ME, McHugh JB, Wolf GT, Rozek LS, Sartor MA. Subtypes of HPV-positive head and neck cancers are associated with HPV characteristics, copy number alterations, PIK3CA mutation, and pathway signatures. Clin Cancer Res. 2016;22(18):4735–45.
• Zhang W, Edwards A, Fang Z, Flemington EK, Zhang K. Integrative genomics and transcriptomics analysis reveals potential mechanisms for favorable prognosis of patients with HPV-positive head and neck carcinomas. Scientific Reports. 2016;6:24927. Exploiting data from TCGA, the authors elucidated the biology of HPV positive HNSCC
Liu Z, Niu Y, Li C, Yang Y, Gao C. Integrating multiple microarray datasets on oral squamous cell carcinoma to reveal dysregulated networks. Head Neck. 2012;34(12):1789–97.
Yang X, Regan K, Huang Y, Zhang Q, Li J, Seiwert TY, Cohen EE, Xing HR, Lussier YA. Single sample expression-anchored mechanisms predict survival in head and neck cancer. PLoS Comput Biol. 2012;8(1):e1002350.
• Sun Y, Sang Z, Jiang Q, Ding X, Yu Y. Transcriptomic characterization of differential gene expression in oral squamous cell carcinoma: a meta-analysis of publicly available microarray data sets. Tumour Biol. 2016 4. Meta-analysis to define potential diagnostic biomarkers and therapeutic targets for OSCC.
Reis PP, Waldron L, Perez-Ordonez B, Pintilie M, Galloni NN, Xuan Y, Cervigne NK, Warner GC, Makitie AA, Simpson C, Goldstein D, Brown D, Gilbert R, Gullane P, Irish J, Jurisica I, Kamel-Reid S. A gene signature in histologically normal surgical margins is predictive of oral carcinoma recurrence. BMC Cancer. 2011;11:437.
Buffa FM, Harris AL, West CM, Miller CJ. Large meta-analysis of multiple cancers reveals a common, compact and highly prognostic hypoxia metagene. Br J Cancer. 2010;102(2):428–35.
•• De Cecco L, Bossi P, Locati L, Canevari S, Licitra L. Comprehensive gene expression meta-analysis of head and neck squamous cell carcinoma microarray data defines a robust survival predictor. Ann Oncol. 2014;25(8):1628–35. Based on meta-analysis approach a prognostic signature was developed and compared to other gene-expression models
Reddy RB, Bhat AR, James BL, Govindan SV, Mathew R, Ravindra DR, Hedne N, Illiayaraja J, Kekatpure V, Khora SS, Hicks W, Tata P, Kuriakose MA, Suresh A. Meta-analyses of microarray datasets identifies ANO1 and FADD as prognostic markers of head and neck cancer. PLoS One. 2016;11(1):e0147409.
Ginos MA, Page GP, Michalowicz BS, Patel KJ, Volker SE, Pambuccian SE, Ondrey FG, Adams GL, Gaffney PM. Identification of a gene expression signature associated with recurrent disease in squamous cell carcinoma of the head and neck. Cancer Res. 2004;64(1):55–63.
Schmalbach CE, Chepeha DB, Giordano TJ, Rubin MA, Teknos TN, Bradford CR, Wolf GT, Kuick R, Misek DE, Trask DK, Hanash S. Molecular profiling and the identification of genes associated with metastatic oral cavity/pharynx squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2004;130(3):295–302.
Warner GC, Reis PP, Jurisica I, Sultan M, Arora S, Macmillan C, Makitie AA, Grénman R, Reid N, Sukhai M, Freeman J, Gullane P, Irish J, Kamel-Reid S. Molecular classification of oral cancer by cDNA microarrays identifies overexpressed genes correlated with nodal metastasis. Int J Cancer. 2004;110(6):857–68.
Roepman P, Wessels LF, Kettelarij N, Kemmeren P, Miles AJ, Lijnzaad P, Tilanus MG, Koole R, Hordijk GJ, van der Vliet PC, Reinders MJ, Slootweg PJ, Holstege FC. An expression profile for diagnosis of lymph node metastases from primary head and neck squamous cell carcinomas. Nat Genet. 2005;37(2):182–6.
Roepman P, Kemmeren P, Wessels LF, Slootweg PJ, Holstege FC. Multiple robust signatures for detecting lymph node metastasis in head and neck cancer. Cancer Res. 2006;66(4):2361–6.
Carinci F, Arcelli D, Lo Muzio L, Francioso F, Valentini D, Evangelisti R, Volinia S, D'Angelo A, Meroni G, Zollo M, Pastore A, Ionna F, Mastrangelo F, Conti P, Tetè S. Molecular classification of nodal metastasis in primary larynx squamous cell carcinoma. Transl Res. 2007;150(4):233–45.
Nguyen ST, Hasegawa S, Tsuda H, Tomioka H, Ushijima M, Noda M, Omura K, Miki Y. Identification of a predictive gene expression signature of cervical lymph node metastasis in oral squamous cell carcinoma. Cancer Sci. 2007;98(5):740–6.
Colella S, Richards KL, Bachinski LL, Baggerly KA, Tsavachidis S, Lang JC, Schuller DE, Krahe R. Molecular signatures of metastasis in head and neck cancer. Head Neck. 2008;30(10):1273–83.
Hensen EF, De Herdt MJ, Goeman JJ, Oosting J, Smit VT, Cornelisse CJ, Baatenburg de Jong RJ. Gene-expression of metastasized versus non-metastasized primary head and neck squamous cell carcinomas: a pathway-based analysis. BMC Cancer. 2008;8:168.
Rickman DS, Millon R, De Reynies A, Thomas E, Wasylyk C, Muller D, Abecassis J, Wasylyk B. Prediction of future metastasis and molecular characterization of head and neck squamous-cell carcinoma based on transcriptome and genome analysis by microarrays. Oncogene. 2008;27(51):6607–22.
Colo AE, Simoes AC, Carvalho AL, Melo CM, Fahham L, Kowalski LP, Soares FA, Neves EJ, Reis LF, Carvalho AF. Functional microarray analysis suggests repressed cell-cell signaling and cell survival-related modules inhibit progression of head and neck squamous cell carcinoma. BMC Med Genet. 2011;4:33.
Van Hooff SR, Leusink FK, Roepman P, Baatenburg de Jong RJ, Speel EJ, van den Brekel MW, van Velthuysen ML, van Diest PJ, van Es RJ, Merkx MA, Kummer JA, Leemans CR, Schuuring E, Langendijk JA, Lacko M, De Herdt MJ, Jansen JC, Brakenhoff RH, Slootweg PJ, Takes RP, Holstege FC. Validation of a gene expression signature for assessment of lymph node metastasis in oral squamous cell carcinoma. J Clin Oncol. 2012;30(33):4104–10.
Jung AC, Job S, Ledrappier S, Macabre C, Abecassis J, de Reyniès A, Wasylyk B. A poor prognosis subtype of HNSCC is consistently observed across methylome, transcriptome, and miRNome analysis. Clin Cancer Res. 2013;19(15):4174–84.
Wang W, Lim WK, Leong HS, Chong FT, Lim TK, Tan DS, Teh BT, Iyer NG. An eleven gene molecular signature for extra-capsular spread in oral squamous cell carcinoma serves as a prognosticator of outcome in patients without nodal metastases. Oral Oncol. 2015;51(4):355–62.
•• Chung CH, Parker JS, Karaca G, Wu J, Funkhouser WK, Moore D, Butterfoss D, Xiang D, Zanation A, Yin X, Shockley WW, Weissler MC, Dressler LG, Shores CG, Yarbrough WG, Perou CM. Molecular classification of head and neck squamous cell carcinomas using patterns of gene expression. Cancer Cell. 2004;5(5):489–500. First study that clearly identified subtypes in HNSCC. The main features of this stratification including clusters related to xenobiotic exposure and mesenchymal pathway was confirmed in other studies
Walter V, Yin X, Wilkerson MD, Cabanski CR, Zhao N, Du Y, Ang MK, Hayward MC, Salazar AH, Hoadley KA, Fritchie K, Sailey CJ, Weissler MC, Shockley WW, Zanation AM, Hackman T, Thorne LB, Funkhouser WD, Muldrew KL, Olshan AF, Randell SH, Wright FA, Shores CG, Hayes DN. Molecular subtypes in head and neck cancer exhibit distinct patterns of chromosomal gain and loss of canonical cancer genes. PLoS One. 2013;8(2):e56823.
•• De Cecco L, Nicolau M, Giannoccaro M, Daidone MG, Bossi P, Locati L, Licitra L, Canevari S. Head and neck cancer subtypes with biological and clinical relevance: meta-analysis of gene-expression data. Oncotarget. 2015;6(11):9627–42. This study represents the larger meta-analysis in HNSCC. Six subtypes were identified including immune-related subtypes
• Keck MK, Zuo Z, Khattri A, Stricker TP, Brown CD, Imanguli M, Rieke D, Endhardt K, Fang P, Brägelmann J, DeBoer R, El-Dinali M, Aktolga S, Lei Z, Tan P, Rozen SG, Salgia R, Weichselbaum RR, Lingen MW, Story MD, Ang KK, Cohen EE, White KP, Vokes EE, Seiwert TY. Integrative analysis of head and neck cancer identifies two biologically distinct HPV and three non-HPV subtypes. Clin Cancer Res. 2015;21(4):870–81. Based on a large cohort of HPV positive tumors, two subtypes of HPV-positive tumors were found
Chung CH, Parker JS, Ely K, Carter J, Yi Y, Murphy BA, Ang KK, El-Naggar AK, Zanation AM, Cmelak AJ, Levy S, Slebos RJ, Yarbrough WG. Gene expression profiles identify epithelial-to-mesenchymal transition andactivation of nuclear factor -kappaB signaling as characteristics of a high-riskhead and neck squamous cell carcinoma. Cancer Res. 2006;66(16):8210–8.
Ganly I, Talbot S, Carlson D, Viale A, Maghami E, Osman I, Sherman E, Pfister D, Chuai S, Shaha AR, Kraus D, Shah JP, Socci ND, Singh B. Identification of angiogenesis/metastases genes predicting chemoradiotherapy response in patients with laryngopharyngeal carcinoma. J Clin Oncol. 2007;25(11):1369–76.
Pramana J, Van den Brekel MW, van Velthuysen ML, Wessels LF, Nuyten DS, Hofland I, Atsma D, Pimentel N, Hoebers FJ, Rasch CR, Begg AC. Gene expression profiling to predict outcome after chemoradiation in head and neck cancer. Int J Radiat Oncol Biol Phys. 2007;69(5):1544–52.
Méndez E, Houck JR, Doody DR, Fan W, Lohavanichbutr P, Rue TC, Yueh B, Futran ND, Upton MP, Farwell DG, Heagerty PJ, Zhao LP, Schwartz SM, Chen C. A genetic expression profile associated with oral cancer identifies a group of patients at high risk of poor survival. Clin Cancer Res. 2009;15(4):1353–61.
de Jong MC, Pramana J, Knegjens JL, Balm AJ, van den Brekel MW, Hauptmann M, Begg AC, Rasch CR. HPV and high-risk gene expression profiles predict response to chemoradiotherapy in head and neck cancer, independent of clinical factors. Radiother Oncol. 2010;95(3):365–70.
Thurlow JK, Peña Murillo CL, Hunter KD, Buffa FM, Patiar S, Betts G, West CM, Harris AL, Parkinson EK, Harrison PR, Ozanne BW, Partridge M, Kalna G. Spectral clustering of microarray data elucidates the roles of microenvironment remodeling and immune responses in survival of head and neck squamous cell carcinoma. J Clin Oncol. 2010;28(17):2881–8.
Fountzilas E, Markou K, Vlachtsis K, Nikolaou A, Arapantoni-Dadioti P, Ntoula E, Tassopoulos G, Bobos M, Konstantinopoulos P, Fountzilas G, Spentzos D. Identification and validation of gene expression models that predict clinical outcome in patients with early-stage laryngeal cancer. Ann Oncol. 2012;23(8):2146–53.
Pavón MA, Parreño M, Téllez-Gabriel M, Sancho FJ, López M, Céspedes MV, Casanova I, Lopez-Pousa A, Mangues MA, Quer M, Barnadas A, León X, Mangues R. Gene expression signatures and molecular markers associated with clinical outcome in locally advanced head and neck carcinoma. Carcinogenesis. 2012;33(9):1707–16.
Rong SN, Xiaohui S, Xiaoyun Q, Chenjie Y, Haiyan W, Xia G. Detection of differentially expressed genes and association with clinicopathological features in laryngeal squamous cell carcinoma. Oncol Lett. 2012;4(6):1354–60.
Fountzilas E, Kotoula V, Angouridakis N, Karasmanis I, Wirtz RM, Eleftheraki AG, Veltrup E, Markou K, Nikolaou A, Pectasides D, Fountzilas G. Identification and validation of a multigene predictor of recurrence in primary laryngeal cancer. PLoS One. 2013;8(8):e70429.
Lohavanichbutr P, Méndez E, Holsinger FC, Rue TC, Zhang Y, Houck J, Upton MP, Futran N, Schwartz SM, Wang P, Chen C. A 13-gene signature prognostic of HPV-negative OSCC: discovery and external validation. Clin Cancer Res. 2013;19(5):1197–203.
• Winter SC, Buffa FM, Silva P, Miller C, Valentine HR, Turley H, Shah KA, Cox GJ, Corbridge RJ, Homer JJ, Musgrove B, Slevin N, Sloan P, Price P, West CM, Harris AL. Relation of a hypoxia metagene derived from head and neck cancer to prognosis of multiple cancers. Cancer Res. 2007;67(7):3441–9. The present work clearly demonstrated the importance of hypoxia signature in the biology of HNSCC
Tomkiewicz C, Hans S, Mucchielli MH, Agier N, Delacroix H, Marisa L, Brasnu D, Aggerbeck LP, Badoual C, Barouki R, Aggerbeck M. A head and neck cancer tumor response-specific gene signature for cisplatin, 5-fluorouracil induction chemotherapy fails with added taxanes. PLoS One. 2012;7(10):e47170.
Chung CH, Lee JW, Slebos RJ, Howard JD, Perez J, Kang H, Fertig EJ, Considine M, Gilbert J, Murphy BA, Nallur S, Paranjape T, Jordan RC, Garcia J, Burtness B, Forastiere AA, Weidhaas JB. A 3'-UTR KRAS-variant is associated with cisplatin resistance in patients with recurrent and/or metastatic head and neck squamous cell carcinoma. Ann Oncol. 2014;25(11):2230–6.
• Bossi P, Bergamini C, Siano M, Cossu Rocca M, Sponghini AP, Favales F, et al. Functional genomics uncover the biology behind the responsiveness of head and neck squamous cell cancer patients to cetuximab. Clin Cancer Res. 2016;22(15):3961–70. First gene-expression analysis proving the idea those expression predictive signatures may be identified in predicting target therapy response
Larsen SR, Johansen J, Sørensen JA, Krogdahl A. The prognostic significance of histological features in oral squamous cell carcinoma. J Oral Pathol Med. 2009;38(8):657–62.
Eustace A, Mani N, Span PN, Irlam JJ, Taylor J, Betts GN, Denley H, Miller CJ, Homer JJ, Rojas AM, Hoskin PJ, Buffa FM, Harris AL, Kaanders JH, West CM. A 26-gene hypoxia signature predicts benefit from hypoxia-modifying therapy in laryngeal cancer but not bladder cancer. Clin Cancer Res. 2013;19(17):4879–88.
Toustrup K, Sørensen BS, Nordsmark M, Busk M, Wiuf C, Alsner J, Overgaard J. Development of a hypoxia gene expression classifier with predictive impact for hypoxic modification of radiotherapy in head and neck cancer. Cancer Res. 2011;71(17):5923–31.
Toustrup K, Sørensen BS, Lassen P, Wiuf C, Alsner J, Overgaard J, Danish Head and Neck Cancer Group (DAHANCA). Gene expression classifier predicts for hypoxic modification of radiotherapy with nimorazole in squamous cell carcinomas of the head and neck. Radiother Oncol. 2012;102(1):122–9.
Eschrich SA, Pramana J, Zhang H, Zhao H, Boulware D, Lee JH, Bloom G, Rocha-Lima C, Kelley S, Calvin DP, Yeatman TJ, Begg AC, Torres-Roca JF. A gene expression model of intrinsic tumor radiosensitivity: prediction of response and prognosis after chemoradiation. Int J Radiat Oncol Biol Phys. 2009;75(2):489–96.
Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N Engl J Med. 2008;359:1116–27.
Seiwert TY, Burtness B, Mehra R, Weiss J, Berger R, Eder JP, Heath K, McClanahan T, Lunceford J, Gause C, Cheng JD, Chow LQ. Safety and clinical activity of pembrolizumab for treatment of recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-012): an open-label, multicentre, phase 1b trial. Lancet Oncol. 2016;17(7):956–65.
Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014. 5.
Acknowledgements
We are grateful to Drs Paolo Bossi e MariaLuisa Sensi for their critical reading of the manuscript and their helpful comments. We would like also to thank all the cited authors for their informative papers that enabled us to organize this review and we apologize for potential missing citations. This work was partially supported by Associazione Italiana Ricerca Cancro (AIRC IG 14750 to SC and AIRC IG 18519 to LDC) and by H2020-EU.3.1. - SOCIETAL CHALLENGES - Health, demographic change and well-being (“BD2Decide; Big Data and models for personalized Head and Neck Cancer Decision support” Project ID: 689715).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Head and Neck Cancer
Luca Tonella and Marco Giannoccaro equally contributing first author
Rights and permissions
About this article
Cite this article
Tonella, L., Giannoccaro, M., Alfieri, S. et al. Gene Expression Signatures for Head and Neck Cancer Patient Stratification: Are Results Ready for Clinical Application?. Curr. Treat. Options in Oncol. 18, 32 (2017). https://doi.org/10.1007/s11864-017-0472-2
Published:
DOI: https://doi.org/10.1007/s11864-017-0472-2