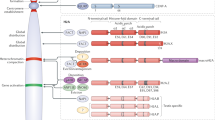
Abstract
Among various histones, histone H1 proteins have been appreciated for their multiple functions in diverse biological processes. In addition to being a structural protein in chromatin, H1 proteins also play critical roles in cell cycle, gene expression, and development. Recent studies reveal the possible effects of H1 in some diseases, such as cancer and neurodegenerative diseases. Here, we review different variants of H1, the functions, and post translational modifications of H1 variants are also discussed.
Similar content being viewed by others
References
Watson J D. The Structure of DNA, in Cold Spring Harbor Symposia on Quantitative Biology [R]. New York: Cold Spring Harbor Laboratory Press, 1953.
Catez F, Ueda T, Bustin M. Determinants of histone H1 mobility and chromatin binding in living cells [J]. Nat Struct Mol Biol, 2006, 13: 305–310.
Zhai Z H, Wang X Z. Cell Biology. 3rd ed [M]. Beijing: Higher Education Press, 2008(Ch).
Tanaka H, Katoh A, Oguro K, et al. Disturbance of hippocampal long-term potentiation after transient ischemia in GFAP deficient mice [J]. J Neurosci Res, 2002, 67: 11–20.
Tanaka M, Hennebold J D, Macfarlane J, et al. A mammalian oocyte-specific linker histone gene H1oo: homology with the genes for the oocyte-specific cleavage stage histone (cs-H1) of sea urchin and the B4/H1M histone of the frog [J]. Development, 2001, 128: 655–664.
Helliger W, Lindner H, Grubl-Knosp O, et al. Alteration in proportions of histone H1 variants during the differentiation of murine erythroleukaemic cells [J]. Biochem J, 1992, 288 (Pt 3): 747–751.
Medrzycki M, Zhang Y, Cao K, et al. Expression analysis of mammalian linker-histone subtypes [EB/OL]. [2013-02-09]. http://www.ncbi.nlm.nih.gov/pubmed/22453355?dopt=Citation.
Medrzycki M, Zhang Y, McDonald J F, et al. Profiling of linker histone variants in ovarian cancer [J]. Front Biosci, 2012, 17: 396–406.
Zhang Y, Cooke M, Panjwani S, et al. Histone h1 depletion impairs embryonic stem cell differentiation [EB/OL]. [2013-05-01]. http://www.ncbi.nlm.nih.gov/pubmed/22589736?dopt=Citation.
Zhang Y, Liu Z, Medrzycki M, et al. Reduction of Hox gene expression by histone H1 depletion [EB/OL]. [2013-06-12]. http://www.ncbi.nlm.nih.gov/pubmed/22701719?dopt=Citation.
Luger K, Mader A W, Richmond R K, et al. Crystal structure of the nucleosome core particle at 2.8 A resolution [J]. Nature, 1997, 389: 251–260.
Richmond T J, Davey C A. The structure of DNA in the nucleosome core [J]. Nature, 2003, 423: 145–150.
Burlingame R W, Love W E, Wang B C, et al. Crystallographic structure of the octameric histone core of the nucleosome at a resolution of 3.3 A [J]. Science, 1985, 228: 546–553.
Du Preez L L, Patterton H G. Secondary structures of the core histone N-terminal tails: Their role in regulating chromatin structure [J]. Subcell Biochem, 2012, 61: 37–55.
Kouzarides T. Chromatin modifications and their function [J]. Cell, 2007, 128: 693–705.
Nightingale K P, O’Neill L P, Turner B M. Histone modifications: Signalling receptors and potential elements of a heritable epigenetic code [J]. Curr Opin Genet Dev, 2006, 16: 125–136.
Galasinski S C, Louie D F, Gloor K K, et al. Global regulation of post-translational modifications on core histones [J]. J Biol Chem, 2002, 277: 2579–2588.
Krishnan S, Horowitz S, Trievel R C. Structure and function of histone H3 lysine 9 methyltransferases and demethylases [J]. Chembiochem, 2011, 12: 254–263.
Kamakaka R T, Biggins S. Histone variants: Deviants? [J]. Genes Dev, 2005, 19: 295–310.
Taty-Taty G C, Courilleau C, Quaranta M, et al. H2A.Z depletion impairs proliferation and viability but not DNA double-strand breaks repair in human immortalized and tumoral cell lines [EB/OL]. [2013-03-09]. https://www.landesbioscience.com/journals/cc/article/27143/.
Hu G, Cui K, Northrup D, et al. H2A.Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation [J]. Cell Stem Cell, 2013, 12: 180–192.
Luo J, Xu X, Hall H, et al. Histone h3 exerts a key function in mitotic checkpoint control [J]. Mol Cell Biol, 2010, 30: 537–549.
Harshman S W, Young N L, Parthun M R, et al. H1 histones: Current perspectives and challenges [J]. Nucleic Acids Res, 2013, 41(21): 9593–9609.
Khochbin S. Histone H1 diversity: bridging regulatory signals to linker histone function [J]. Gene, 2001, 271: 1–12.
Bustin M, Catez F, Lim J H. The dynamics of histone H1 function in chromatin [J]. Mol Cell, 2005, 17: 617–620.
Sancho M, Diani E, Beato M, et al. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth [EB/OL]. [2012-11-20]. http://www.ncbi.nlm. nih.gov/pubmed/18927631?dopt=Citation.
Abel T W, Clark C, Bierie B, et al. GFAP-Cre-mediated activation of oncogenic K-ras results in expansion of the subventricular zone and infiltrating glioma [J]. Mol Cancer Res, 2009, 7: 645–653.
Bernal G M, Peterson D A. Phenotypic and gene expression modification with normal brain aging in GFAP-positive astrocytes and neural stem cells [J]. Aging Cell, 2011, 10: 466–482.
Meergans T, Albig W, Doenecke D. Varied expression patterns of human H1 histone genes in different cell lines [J]. DNA Cell Biol, 1997, 16: 1041–1049.
Terme J M, Sese B, Millan-Arino L, et al. Histone H1 variants are differentially expressed and incorporated into chromatin during differentiation and reprogramming to pluripotency [J]. J Biol Chem, 2011, 286: 35347–35357.
Izzo A, Kamieniarz K, Schneider R. The histone H1 family: Specific members, specific functions? [J]. Biol Chem, 2008, 389: 333–343.
McBryant S J, Lu X, Hansen J C. Multifunctionality of the linker histones: an emerging role for protein-protein interactions [J]. Cell Res, 2010, 20: 519–528.
Babu M, Bai R P, Suguna L, et al. Differentiation of keloid and hypertrophic scar; Correlation of the water proton relaxation times with the duration of the scar [J]. Physiol Chem Phys Med NMR, 1993, 25: 113–120.
Cerf C, Lippens G, Ramakrishnan V, et al. Homo- and heteronuclear two-dimensional NMR studies of the globular domain of histone H1: Full assignment, tertiary structure, and comparison with the globular domain of histone H5 [J]. Biochemistry, 1994, 33: 11079–11086.
Clark K L, Halay E D, Lai E, et al. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5 [J]. Nature, 1993, 364: 412–420.
Kasinsky H E, Lewis J D, Dacks J B, et al. Origin of H1 linker histones [J]. FASEB J, 2001, 15: 34–42.
Hendzel M J, Lever M A, Crawford E, et al. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo [J]. J Biol Chem, 2004, 279: 20028–20034.
Kalashnikova A A, Winkler D D, McBryant S J, et al. Linker histone H1.0 interacts with an extensive network of proteins found in the nucleolus [J]. Nucleic Acids Res, 2013, 41: 4026–4035.
Orrego M, Ponte I, Roque A, et al. Differential affinity of mammalian histone H1 somatic subtypes for DNA and chromatin [J]. BMC Biol, 2007, 5: 1–11.
Th’ng J P, Sung R, Ye M, et al. H1 family histones in the nucleus. Control of binding and localization by the C-terminal domain [J]. J Biol Chem, 2005, 280: 27809–27814.
Clausell J, Happel N, Hale T K, et al. Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF [EB/OL]. [2012-10-07]. http://www.ncbi.nlm.nih.gov/pubmed/19794910?dopt=Citation.
Balhorn R, Chalkley R, Granner D. Lysine-rich histone phosphorylation. A positive correlation with cell replication [J]. Biochemistry, 1972, 11: 1094–1098.
Garcia B A, Busby S A, Barber C M, et al. Characterization of phosphorylation sites on histone H1 isoforms by tandem mass spectrometry [J]. J Proteome Res, 2004, 3: 1219–1227.
Wisniewski J R, Zougman A, Kruger S, et al. Mass spectrometric mapping of linker histone H1 variants reveals multiple acetylations, methylations, and phosphorylation as well as differences between cell culture and tissue [J]. Mol Cell Proteomics, 2007, 6: 72–87.
Ruan J, Ouyang H, Amaya M F, et al. Structural basis of the chromodomain of Cbx3 bound to methylated peptides from histone h1 and G9a [EB/OL]. [2013-04-21].http://www.ncbi.nlm.nih.gov/pubmed/22514736?dopt=Citation.
Bonet-Costa C, Vilaseca M, Diema C, et al. Combined bottom-up and top-down mass spectrometry analyses of the pattern of post-translational modifications of Drosophila melanogaster linker histone H1 [J]. J Proteomics, 2012, 75: 4124–4138.
Lu A, Zougman A, Pudelko M, et al. Mapping of lysine monomethylation of linker histones in human breast and its cancer [J]. J Proteome Res, 2009, 8: 4207–4215.
Bruce A, Alexander J, Julian L, et al. Molecular Biology of THE CELL [M]. New York: Garland Science Press, 2007: 1392.
Ruiz-Carrillo A, Puigdomenech P, Eder G, et al. Stability and reversibility of higher ordered structure of interphase chromatin: continuity of deoxyribonucleic acid is not required for maintenance of folded structure [J]. Biochemistry, 1980, 19: 2544–2554.
Hansen J C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions [J]. Annu Rev Biophys Biomol Struct, 2002, 31: 361–392.
Shen X, Yu L, Weir J W, et al. Linker histones are not essential and affect chromatin condensation in vivo [J]. Cell, 1995, 82: 47–56.
Thoma F, Koller T, Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin [J]. J Cell Biol, 1979, 83: 403–427.
Simpson R T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones [J]. Biochemistry, 1978, 17: 5524–5531.
Renz M, Nehls P, Hozier J. Involvement of histone H1 in the organization of the chromosome fiber [J]. Proc Natl Acad Sci U S A, 1977, 74: 1879–1883.
Green G R, Lee H J, Poccia D L. Phosphorylation weakens DNA binding by peptides containing multiple “SPKK” sequences [J]. J Biol Chem, 1993, 268: 11247–11255.
Kassner I, Barandun M, Fey M, et al. Crosstalk between SET7/9-dependent methylation and ARTD1-mediated ADP-ribosylation of histone H1.4 [J]. Epigenetics Chromatin, 2013, 6: 1–9.
Kowalski A, Palyga J. Linker histone subtypes and their allelic variants [J]. Cell Biol Int, 2012, 36: 981–996.
Brown D T, Alexander B T, Sittman D B. Differential effect of H1 variant overexpression on cell cycle progression and gene expression [J]. Nucleic Acids Res, 1996, 24: 486–493.
Croston G E, Kerrigan L A, Lira L M, et al. Sequence-specific antirepression of histone H1-mediated inhibition of basal RNA polymerase II transcription [J]. Science, 1991, 251: 643–649.
Laybourn P J, Kadonaga J T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II [J]. Science, 1991, 254: 238–245.
Gao B, Jaffe H, Kunos G. Histone H1 isoforms purified from rat liver bind nonspecifically to the nuclear factor 1 recognition sequence and serve as generalized transcriptional repressors [J]. Mol Cell Biochem, 1998, 178: 187–196.
Abdelfadil E, Cheng Y H, Bau D T, et al. Thymoquinone induces apoptosis in oral cancer cells through p38beta inhibition [J]. Am J Chin Med, 2013, 41: 683–696.
Anh T D, Ahn M Y, Kim S A, et al. The histone deacetylase inhibitor, Trichostatin A, induces G2/M phase arrest and apoptosis in YD-10B oral squamous carcinoma cells [J]. Oncol Rep, 2012, 27: 455–460.
Messing A, Li R, Naidu S, et al. Error in Figure in: Archetypal and New Families With lexander Disease and Novel Mutations in GFAP [J]. Arch Neurol, 2012, 69: 208–214.
Hellauer K, Sirard E, Turcotte B. Decreased expression of specific genes in yeast cells lacking histone H1 [J]. J Biol Chem, 2001, 276: 13587–13592.
Karrer K M, Peiffer S L, DiTomas M E. Two distinct gene subfamilies within the family of cysteine protease genes [J]. Proc Natl Acad Sci U S A, 1993, 90: 3063–3067.
Shen X, Gorovsky M A. Linker histone H1 regulates specific gene expression but not global transcription in vivo [J]. Cell, 1996, 86: 475–483.
Mori N, Matsuda T, Tadano M, et al. Apoptosis induced by the histone deacetylase inhibitor FR901228 in human T-cell leukemia virus type 1-infected T-cell lines and primary adult T-cell leukemia cells. [J]. J Virol, 2011, 85: 1414–1415.
Kamieniarz K, Izzo A, Dundr M, et al. A dual role of linker histone H1.4 Lys 34 acetylation in transcriptional activation [J]. Genes Dev, 2012, 26: 797–802.
Herrera R E, Chen F, Weinberg R A. Increased histone H1 phosphorylation and relaxed chromatin structure in Rb-deficient fibroblasts [J]. Proc Natl Acad Sci U S A, 1996, 93: 11510–11515.
Bradbury E M. Reversible histone modifications and the chromosome cell cycle [J]. Bioessays, 1992, 14: 9–16.
Hohmann P, Tobey R A, Gurley L R. Phosphorylation of distinct regions of f1 histone. Relationship to the cell cycle [J]. J Biol Chem, 1976, 251: 3685–3692.
Chu C S, Hsu P H, Lo P W, et al. Protein kinase A-mediated serine 35 phosphorylation dissociates histone H1.4 from mitotic chromosome [J]. J Biol Chem, 2011, 286: 35843–35851.
Woodcock C L, Skoultchi A I, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length [J]. Chromosome Res, 2006, 14: 17–25.
Dworkin-Rastl E, Kandolf H, Smith R C. The maternal histone H1 variant, H1M (B4 protein), is the predominant H1 histone in Xenopus pregastrula embryos [J]. Dev Biol, 1994, 161: 425–439.
Fan Y, Sirotkin A, Russell R G, et al. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype [J]. Mol Cell Biol, 2001, 21: 7933–7943.
Fan Y, Nikitina T, Morin-Kensicki E M, et al. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo [J]. Mol Cell Biol, 2003, 23: 4559–4572.
Konishi A, Shimizu S, Hirota J, et al. Involvement of histone H1.2 in apoptosis induced by DNA double-strand breaks [J]. Cell, 2003, 114: 673–688.
Awasthi S, Singhal S S, Singhal J, et al. Role of RLIP76 in lung cancer doxorubicin resistance: III. Anti-RLIP76 antibodies trigger apoptosis in lung cancer cells and synergistically increase doxorubicin cytotoxicity [J]. Int J Oncol, 2003, 22: 721–732.
Vadlamudi R K, Wang R A, Mazumdar A, et al. Molecular cloning and characterization of PELP1, a novel human coregulator of estrogen receptor alpha [J]. J Biol Chem, 2001, 276: 38272–38279.
Nair S S, Mishra S K, Yang Z, et al. Potential role of a novel transcriptional coactivator PELP1 in histone H1 displacement in cancer cells [J]. Cancer Res, 2004, 64: 6416–6423.
Jani A B, Hellman S. Early prostate cancer: Elinical decision-making [J]. Lancet, 2003, 361: 1045–1053.
Merglen A, Schmidlin F, Fioretta G, et al. Short- and long-term mortality with localized prostate cancer [J]. Arch Intern Med, 2007, 167: 1944–1950.
Dotiwala F, Eapen V V, Harrison J C, et al. DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase [J]. Proc Natl Acad Sci U S A, 2013, 110: 41–49.
Makin O S, Serpell L C. Structures for amyloid fibrils [J]. FEBS J, 2005, 272: 5950–5961.
Westermark P, Benson M D, Buxbaum J N, et al. Amyloid: Toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis [J]. Amyloid, 2005, 12: 1–4.
Gordon D J, Meredith S C. Probing the role of backbone hydrogen bonding in beta-amyloid fibrils with inhibitor peptides containing ester bonds at alternate positions [J]. Biochemistry, 2003, 42: 475–485.
Duce J A, Smith D P, Blake R E, et al. Linker histone H1 binds to disease associated amyloid-like fibrils [J]. J Mol Biol, 2006, 361: 493–505.
Yamada M, Sato T, Tsuji S, et al. CAG repeat disorder models and human neuropathology: similarities and differences [J]. Acta Neuropathol, 2008, 115: 71–86.
Durr A. Autosomal dominant cerebellar ataxias: polyglutamine expansions and beyond [J]. Lancet Neurol, 2010, 9: 885–894.
Cummings C J, Zoghbi H Y. Trinucleotide repeats: Mechanisms and pathophysiology [J]. Annu Rev Genomics Hum Genet, 2000, 1: 281–328.
Kizilyaprak C, Spehner D, Devys D, et al. The linker histone H1C contributes to the SCA7 nuclear phenotype [J]. Nucleus, 2011, 2: 444–454.
Misteli T, Gunjan A, Hock R, et al. Dynamic binding of histone H1 to chromatin in living cells [J]. Nature, 2000, 408: 877–881.
Lever M A, Th’ng J P, Sun X, et al. Rapid exchange of histone H1.1 on chromatin in living human cells [J]. Nature, 2000, 408: 873–876.
Arar N H, Freedman B I, Adler S G, et al. Heritability of the severity of diabetic retinopathy: the FIND-Eye study [J]. Invest Ophthalmol Vis Sci, 2008, 49: 3839–3845.
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: Supported by the National Basic Research Program of China (2012CB524901), the Natural Science Foundation of China (31271370, 81100687), and the Program for New Century Excellent Talents in University (NECT10-0623)
Biography: WANG Wenjun, female, Ph.D. candidate, research direction: diabetes and its complications.
Rights and permissions
About this article
Cite this article
Wang, W., Cai, R., Xiao, H. et al. Multifunctions of histone H1 proteins. Wuhan Univ. J. Nat. Sci. 19, 8–18 (2014). https://doi.org/10.1007/s11859-014-0972-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11859-014-0972-x