Abstract
Background
Defects in neurotransmission and synaptogenesis are noteworthy in the pathogenesis of ASD. Synapsin III (SYN III) is defined as a synaptic vesicle protein that plays an important role in synaptogenesis and regulation of neurotransmitter release and neurite outgrowth. Therefore, SYN III may associate with many neurodevelopmental diseases, including ASD.
Aim
The aim of this study was to investigate whether the SYN III gene -631 C > G (rs133946) and -196 G > A (rs133945) polymorphisms are associated with susceptibility to ASD.
Methods
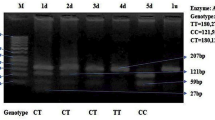
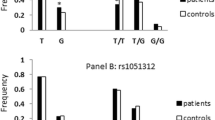
SYN III variants and the risk of ASD were investigated in 26 healthy children and 24 ASD children. SYN III gene variants were genotyped by PCR–RFLP methods. The differences in genotype and allele frequencies between the ASD and control groups were calculated using the chi-square (χ2). We analysed the SYN III gene using web-based tools.
Results
Our results suggest that the presence of the AA genotype of the SYN III -196 G > A (rs133945) polymorphism affects the characteristics and development of ASD in children (p = 0.012). SYN III -631 C > G (rs133946) polymorphism was not associated with ASD (p = 0.524). We have shown the prediction of gene–gene interaction that SYN III is co-expressed with 17 genes, physical interaction with 3 genes, and co-localization with 12 genes. The importance of different genes (SYN I, II, III, GABRD, NOS1AP, GNAO1) for ASD pathogenesis was revealed by GO analysis.
Conclusion
Considering the role of SYN III and related genes, especially in the synaptic vesicle pathway and neurotransmission, its effect on ASD can be further investigated.



Similar content being viewed by others
References
Chen JA, Peñagarikano O, Belgard TG et al (2015) The emerging picture of autism spectrum disorder: genetics and pathology. Annu Rev Pathol 10:111–144
Geschwind DH (2011) Genetics of autism spectrum disorders. Trends Cogn Sci 15(9):409–416
Lord C, Elsabbagh M, Baird G et al (2018) Autism spectrum disorder. Lancet 392(10146):508–520
Ivanov HY, Stoyanova VK, Popov NT et al (2015) Autism spectrum disorder - a complex genetic disorder. Folia Med (Plovdiv) 57(1):19–28
Şahın N, Kara M, Kara B, Topal H (2018) Evaluation of RELN gene polymorphism in children with autism spectrum disorder. Anadolu Psikiyatri Dergisi 19:599–606
Ecker C (2017) The neuroanatomy of autism spectrum disorder: an overview of structural neuroimaging findings and their translatability to the clinical setting. Autism 21(1):18–28
Bourgeron T (2015) From the genetic architecture to synaptic plasticity in autism spectrum disorder. Nat Rev Neurosci 16(9):551–563. https://doi.org/10.1038/nrn3992
Özdemir Ç, Şahin N, Edgünlü T (2022) Vesicle trafficking with snares: a perspective for autism. Mol Biol Rep 49(12):12193–12202
Salpietro V, Malintan NT, Llano-Rivas I et al (2019) Mutations in the neuronal vesicular SNARE VAMP2 affect synaptic membrane fusion and impair human neurodevelopment. Am J Hum Genet 104(4):721–730
Nakamura K, Iwata Y, Anitha A et al (2011) Replication study of Japanese cohorts supports the role of STX1A in autism susceptibility. Prog Neuropsychopharmacol Biol Psychiatry 35(2):454–458
Safari MR, Omrani MD, Noroozi R et al (2017) Synaptosome-associated protein 25 (SNAP25) gene association analysis revealed risk variants for ASD. Iranian population J Mol Neurosci 61(3):305–311
Fujiwara T, Snada M, Kofuji T et al (2010) HPC-1/syntaxin 1A gene knockout mice show abnormal behavior possibly related to a disruption in 5-HTergic systems. Eur J Neurosci 32(1):99–107
Greco B, Managò F, Tucci V et al (2013) Autism-related behavioral abnormalities in synapsin knockout mice. Behav Brain Res 251:65–74. https://doi.org/10.1016/j.bbr.2012.12.015
Chen J, Yu S, Fu Y et al (2014) Synaptic proteins and receptors defects in autism spectrum disorders. Front Cell Neurosci 8:276
Tang BL (2020) SNAREs and developmental disorders. J Cell Physiol 236(4):2482–2504. https://doi.org/10.1002/jcp.30067
Feng J, Chi P, Blanpied TA et al (2002) Regulation of neurotransmitter release by synapsin III. J Neurosci 22(11):4372–4380. https://doi.org/10.1523/JNEUROSCI.22-11-04372.2002
Michetti C, Caruso A, Pagani M et al (2017) The knockout of synapsin II in mice impairs social behavior and functional connectivity generating an ASD-like phenotype. Cereb Cortex 27(10):5014–5023. https://doi.org/10.1093/cercor/bhx207
Lee E, Lee J, Kim E (2017) Excitation/inhibition imbalance in animal models of autism spectrum disorders. Biol Psychiat 81(10):838–847
Alymov AA, Kapitsa IG, Voronina TA (2021) Neurochemical mechanisms of pathogenesis and pharmacological correction of autism spectrum disorders: current concepts and prospects. Neurochem J 15(2):129–138. https://doi.org/10.1134/S1819712421020033
Porton B, Wetsel WC, Kao HT (2011) Synapsin III: role in neuronal plasticity and disease. Semin Cell Dev Biol 22(4):416–424. https://doi.org/10.1016/j.semcdb.2011.07.007
Kenar ANI, Edgünlü T, Herken H et al (2013) Association of synapsin III gene with adult attention deficit hyperactivity disorder. DNA Cell Biol 32(8):430–434
Warde-Farley D, Donaldson SL, Comes O et al (2010) The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38(Web Server issue):W214–W220
Gilani N, Belaghi RA, Aftabi Y et al (2021) Identifying potential miRNA biomarkers for gastric cancer diagnosis using machine learning variable selection approach. Front Genet 12
Kile BM, Guillot TS, Venton BJ et al (2010) Synapsins differentially control dopamine and serotonin release. J Neurosci 30(29):9762–9770
Ferreira A, Kao HT, Feng J et al (2000) Synapsin III: developmental expression, subcellular localization, and role in axon formation. J Neurosci 20(10):3736–3744
Kao HT, Li P, Chao HM et al (2008) Early involvement of synapsin III in neural progenitor cell development in the adult hippocampus. J Comp Neurol 507(6):1860–1870
Ekinci A, Ekinci O, Turkcarpar H et al (2012) Emotional schemas and their relationship with clinical characteristics in patients with alcohol dependence/Alkol bagimlisi olgularin saglikli kontrollerle emosyonel semalar yonunden karsilastirilmasi ve klinik ozelliklerle iliskisi. Noro Psikiyatr Ars 49(4):286–294
Başay Ö, Kabukcu Basay B, Alacam H et al (2016) The impact of synapsin III gene on the neurometabolite level alterations after single-dose methylphenidate in attention-deficit hyperactivity disorder patients. Neuropsychiatr Dis Treat 12:1141–1149
Porton B, Ferreira A, DeLisi LE et al (2004) A rare polymorphism affects a mitogen-activated protein kinase site in synapsin III: possible relationship to schizophrenia. Biol Psychiatry 55(2):118–125
Hall H, Lawyer G, Sillen A et al (2007) Potential genetic variants in schizophrenia: a Bayesian analysis. World J Biol Psychiatry 8(1):12–22
Akkad DA, Gödde R, Epplen JT (2006) No association between synapsin III gene promoter polymorphisms and multiple sclerosis in German patients. J Neurol 253(10):1365–1366
Otaegui D, Zuriarrain O, Castillo-Trivino T et al (2009) Association between synapsin III gene promoter SNPs and multiple sclerosis in Basque patients. Mult Scler 15(1):124–128
Bolat H, Ünsel-Bolat G, Özgül S et al (2022) Investigation of possible associations of the BDNF, SNAP-25 and SYN III genes with the neurocognitive measures: BDNF and SNAP-25 genes might be involved in attention domain, SYN III gene in executive function. Nord J Psychiatry 1–6
Edgünlü TG, Özge A, Yalin OÖ et al (2012) Genetic variants of synaptic vesicle and presynaptic plasma membrane proteins in Alzheimer’s disease. Noro Psikiyatr Ars 49(4)
Giovedí S, Corradi A, Fassio A et al (2014) Involvement of synaptic genes in the pathogenesis of autism spectrum disorders: the case of synapsins. Front Pediatr 2:94
Ahring PK, Liao VW, Gardella E et al (2022) Gain-of-function variants in GABRD reveal a novel pathway for neurodevelopmental disorders and epilepsy. Brain 145(4):1299–1309
Sesarini CV, Costa L, Naymark M et al (2014) Evidence for interaction between markers in GABA (A) receptor subunit genes in an A rgentinean autism spectrum disorder population. Autism Res 7(1):162–166
Lu ATH, Dai X, Martinez-Agosto JA et al (2012) Support for calcium channel gene defects in autism spectrum disorders. Mol autism 3(1):1–9
Delorme R, Betancur C, Scheid I et al (2010) Mutation screening of NOS1AP gene in a large sample of psychiatric patients and controls. BMC Med Genet 11(1):1–9
Hepler JR, Gilman AG (1992) G proteins. Trends Biochem Sci 17(10):383–387
Jiang M, Bajpayee NS (2009) Molecular mechanisms of go signaling. Neurosignals 17(1):23–41
Jiang M, Gold MS, Boulay G et al (1998) Multiple neurological abnormalities in mice deficient in the G protein Go. Proc Natl Acad Sci 95(6):3269–3274
Skafidas E, Testa R, Zantomio D et al (2014) Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Mol Psychiatry 19(4):504–510
Gerald B, Ramsey K, Belnap N et al (2018) Neonatal epileptic encephalopathy caused by de novo GNAO1 mutation misdiagnosed as atypical Rett syndrome: cautions in interpretation of genomic test results. In Semin Pediatr Neurol 26:28–32
Schirinzi T, Garone G, Travaglini L et al (2019) Phenomenology and clinical course of movement disorder in GNAO1 variants: results from an analytical review. Parkinsonism Relat Disord 61:19–25
Danti FR, Galosi S, Romani M et al (2017) GNAO1 encephalopathy: broadening the phenotype and evaluating treatment and outcome. Neurol Genet 3:e143
Muir AM, Myers CT, Nguyen NT et al (2019) Genetic heterogeneity in infantile spasms. Epilepsy Res 156:106181
Marcé-Grau A, Dalton J, López-Pisón J et al (2016) GNAO1 encephalopathy: further delineation of a severe neurodevelopmental syndrome affecting females. Orphanet J Rare Dis 11(1):1–9
Lakhan R, Kalita J, Misra UK et al (2010) Association of intronic polymorphism rs3773364 A> G in synapsin-2 gene with idiopathic epilepsy. Synapse 64(5):403–408
O’Connell KS, McGregor NW, Lochner C et al (2018) The genetic architecture of schizophrenia, bipolar disorder, obsessive-compulsive disorder and autism spectrum disorder. Mol Cell Neurosci 88:300–307
Sharma AR, Batra G, Saini L et al (2022) Valproic acid and propionic acid modulated mechanical pathways associated with autism spectrum disorder at prenatal and neonatal exposure. CNS Neurol Disord Drug Targets 21(5):399–408
Fassio A, Patry L, Congia S et al (2011) SYN1 loss-of-function mutations in autism and partial epilepsy cause impaired synaptic function. Hum Mol Genet 20(12):2297–2307
Funding
This paper was supported by the Scientific and Technological Research Council of Turkey (TUBITAK) under Grant No: 1919B012003527.
Author information
Authors and Affiliations
Contributions
Remzi Oguz BARIS: Investigation, Writing-Original Draft, Visualization; Nilfer SAHİN: Conceptualizaton, Resources, Investigation, Writing-Original Draft, Visualization; Aysegul DEMIRTAS BILGIC: Investigation, Writing-Original Draft, Visualization; Cilem OZDEMİR: Investigation, Writing-Original Draft, Visualization; Tuba GOKDOGAN EDGUNLU: Conceptualizaton, Resources, Writing-Review and Editing, Supervision, Project Administration. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The study was approved by Muğla Sıtkı Koçman University Medical Faculty Ethics Committee and was conducted according to the Declaration of Helsinki, and informed consent was obtained from all participants. The procedures used in this study adhere to the tenets of the Declaration of Helsinki. Informed consent and written informed consent were obtained from all patients prior to their participation in the study.
Consent for publication
Consent has been granted by all authors for the publication of this manuscript.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baris, R.O., Sahin, N., Bilgic, A.D. et al. Molecular and in silico analyses of SYN III gene variants in autism spectrum disorder. Ir J Med Sci 192, 2887–2895 (2023). https://doi.org/10.1007/s11845-023-03402-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-023-03402-w