Abstract
Objective
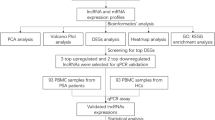
This study aimed to explore the dysregulated long non-coding RNA (lncRNA) expression profile in psoriatic tissue vs. normal skin tissue via RNA sequencing (RNA-seq), then further sort candidate lncRNAs to be validated by reverse transcription quantitative polymerase chain reaction (RT-qPCR), in order to investigate the comprehensive linkage of lncRNA with psoriasis.
Methods
Twenty-five psoriasis patients were consecutively enrolled, with their psoriatic and surrounding normal skin tissues obtained. Ten pairs of psoriatic and normal tissues were proposed to RNA-seq. Then, top 6 differentially expressed lncRNAs (DElncRNA) were sorted as candidate lncRNAs for validation by RT-qPCR in 25 pairs of samples.
Results
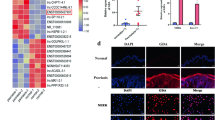
Principal component analysis (PCA) exhibited that lncRNA profile clearly distinguished psoriatic tissue from normal tissue, so did heatmap. Volcano plot disclosed 412 upregulated and 625 downregulated DElncRNAs in psoriatic tissue vs. normal tissue. Gene Ontology (GO) and Kyoko Encyclopedia of Genes and Genomes (KEGG) enrichment analyses exhibited that these DElncRNAs were mainly enriched in immune, inflammation, or proliferation-related biological processes and pathways such as neutrophil degranulation, regulation of immune response, positive regulation of cell proliferation, and MAPK signaling pathway. By RT-qPCR validation, lncRNAs RP11-22A3.2, RP11-342L8.2, and CTD-2006H14.2 were increased (all P < 0.001), while lncRNAs AP000442.4, CCDC144NL-AS1, and MIR663AHG were decreased (all P < 0.01) in psoriatic tissue vs. normal tissue. Interestingly, psoriatic lncRNA RP11-342L8.2 was also observed to positively correlated with psoriasis area and severity index (PASI) (r = 0.405, P = 0.045).
Conclusion
Our present study exhibits some evidence for the landscape of lncRNAs implicated in psoriasis.




Similar content being viewed by others
References
Nair PA, Badri T (2021) Psoriasis. In: StatPearls. Treasure Island (FL)
Vicic M, Kastelan M, Brajac I et al (2021) Current concepts of psoriasis immunopathogenesis. Int J Mol Sci 22(21). https://doi.org/10.3390/ijms222111574
Gamret AC, Price A, Fertig RM et al (2018) Complementary and alternative medicine therapies for psoriasis: a systematic review. JAMA Dermatol 154(11):1330–1337. https://doi.org/10.1001/jamadermatol.2018.2972
Weber B, Merola JF, Husni ME et al (2021) Psoriasis and cardiovascular disease: Novel mechanisms and evolving therapeutics. Curr Atheroscler Rep 23(11):67. https://doi.org/10.1007/s11883-021-00963-y
Rademaker M, Agnew K, Anagnostou N et al (2019) Psoriasis and infection. A clinical practice narrative. Australas J Dermatol 60(2):91–98. https://doi.org/10.1111/ajd.12895
Zabotti A, De Lucia O, Sakellariou G et al (2021) Predictors, risk factors, and incidence rates of psoriatic arthritis development in psoriasis patients: a systematic literature review and meta-analysis. Rheumatol Ther 8(4):1519–1534. https://doi.org/10.1007/s40744-021-00378-w
Sahi FM, Masood A, Danawar NA et al (2020) Association between psoriasis and depression: a traditional review. Cureus 12(8):e9708. https://doi.org/10.7759/cureus.9708
Gooderham M, Gavino-Velasco J, Clifford C et al (2016) A review of psoriasis, therapies, and suicide. J Cutan Med Surg 20(4):293–303. https://doi.org/10.1177/1203475416648323
Pezzolo E, Naldi L (2019) The relationship between smoking, psoriasis and psoriatic arthritis. Expert Rev Clin Immunol 15(1):41–48. https://doi.org/10.1080/1744666X.2019.1543591
Ghafouri-Fard S, Eghtedarian R, Taheri M, Rakhshan A (2020) The eminent roles of ncRNAs in the pathogenesis of psoriasis. Noncoding RNA Res 5(3):99–108. https://doi.org/10.1016/j.ncrna.2020.06.002
Taheri M, Eghtedarian R, Dinger ME, Ghafouri-Fard S (2020) Dysregulation of non-coding RNAs in rheumatoid arthritis. Biomed Pharmacother 130:110617. https://doi.org/10.1016/j.biopha.2020.110617
Huang D, Liu J, Wan L et al (2021) Identification of lncRNAs associated with the pathogenesis of ankylosing spondylitis. BMC Musculoskelet Disord 22(1):272. https://doi.org/10.1186/s12891-021-04119-6
Ghafouri-Fard S, Eghtedarian R, Taheri M (2020) The crucial role of non-coding RNAs in the pathophysiology of inflammatory bowel disease. Biomed Pharmacother 129:110507. https://doi.org/10.1016/j.biopha.2020.110507
Li S, Zhu X, Zhang N et al (2021) LncRNA NORAD engages in psoriasis by binding to miR-26a to regulate keratinocyte proliferation. Autoimmunity 54(3):129–137. https://doi.org/10.1080/08916934.2021.1897976
Jia HY, Zhang K, Lu WJ et al (2019) LncRNA MEG3 influences the proliferation and apoptosis of psoriasis epidermal cells by targeting miR-21/caspase-8. BMC Mol Cell Biol 20(1):46. https://doi.org/10.1186/s12860-019-0229-9
Su F, Jin L, Liu W (2021) MicroRNA-125a correlates with decreased psoriasis severity and inflammation and represses keratinocyte proliferation. Dermatology 237(4):568–578. https://doi.org/10.1159/000510681
Head SR, Komori HK, LaMere SA et al (2014) Library construction for next-generation sequencing: overviews and challenges. Biotechniques 56(2):61–64, 66, 68, passim. https://doi.org/10.2144/000114133
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15(12):550. https://doi.org/10.1186/s13059-014-0550-8
Huang DW, Sherman BT, Tan Q et al (2007) DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res 35(Web Server issue):W169–175. https://doi.org/10.1093/nar/gkm415
Ahmed Shehata W, Maraee A, Abd El Monem Ellaithy M et al (2021) Circulating long noncoding RNA growth arrest-specific transcript 5 as a diagnostic marker and indicator of degree of severity in plaque psoriasis. Int J Dermatol 60(8):973–979. https://doi.org/10.1111/ijd.15494
Elamir AM, Shaker OG, El-Komy MH et al (2021) The role of LncRNA MALAT-1 and MiRNA-9 in psoriasis. Biochem Biophys Rep 26:101030. https://doi.org/10.1016/j.bbrep.2021.101030
Yao X, Hao S, Xue T et al (2021) Association of HOTAIR polymorphisms with susceptibility to psoriasis in a Chinese Han population. Biomed Res Int 2021:5522075. https://doi.org/10.1155/2021/5522075
Yan J, Song J, Qiao M et al (2019) Long noncoding RNA expression profile and functional analysis in psoriasis. Mol Med Rep 19(5):3421–3430. https://doi.org/10.3892/mmr.2019.9993
Kvist-Hansen A, Kaiser H, Wang X et al (2021) Neutrophil pathways of inflammation characterize the blood transcriptomic signature of patients with psoriasis and cardiovascular disease. Int J Mol Sci 22(19). https://doi.org/10.3390/ijms221910818
Tang KW, Lin ZC, Wang PW et al (2020) Facile skin targeting of a thalidomide analog containing benzyl chloride moiety alleviates experimental psoriasis via the suppression of MAPK/NF-kappaB/AP-1 phosphorylation in keratinocytes. J Dermatol Sci 99(2):90–99. https://doi.org/10.1016/j.jdermsci.2020.05.013
Lei H, Li X, Jing B et al (2017) Human S100A7 induces mature interleukin1alpha expression by RAGE-p38 MAPK-Calpain1 pathway in psoriasis. PLoS One 12(1):e0169788. https://doi.org/10.1371/journal.pone.0169788
Zhang L, Chi B, Chai J et al (2021) LncRNA CCDC144NL-AS1 serves as a prognosis biomarker for non-small cell lung cancer and promotes cellular function by targeting miR-490-3p. Mol Biotechnol 63(10):933–940. https://doi.org/10.1007/s12033-021-00351-6
Zhang Y, Zhang H, Wu S (2021) LncRNA-CCDC144NL-AS1 promotes the development of hepatocellular carcinoma by inducing WDR5 expression via sponging miR-940. J Hepatocell Carcinoma 8:333–348. https://doi.org/10.2147/JHC.S306484
Huang J, Li Y, Ye Z et al (2021) Prediction of a potential mechanism of intervertebral disc degeneration based on a novel competitive endogenous RNA network. Biomed Res Int 2021:6618834. https://doi.org/10.1155/2021/6618834
Nakagawa S, Kawashima M, Miyatake Y et al (2021) Expression of ERV3-1 in leukocytes of acute myelogenous leukemia patients. Gene 773:145363. https://doi.org/10.1016/j.gene.2020.145363
Kakuta Y, Ichikawa R, Fuyuno Y et al (2020) An integrated genomic and transcriptomic analysis reveals candidates of susceptibility genes for Crohn’s disease in Japanese populations. Sci Rep 10(1):10236. https://doi.org/10.1038/s41598-020-66951-5
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The ethical approval was obtained from the Institutional Review Board of Tongren Hospital, Shanghai Jiao Tong University School of Medicine.
Consent to participate
All patients signed the informed consents.
Consent to publish
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11845_2021_2882_MOESM4_ESM.tif
Supplementary file4: Bioinformatic analyses of mRNA profile. PCA plot analysis (A) and heatmap analysis (B) of mRNA profile between psoriatic and normal tissues. DEmRNAs exhibited by Volcano plot analysis (C). GO enrichment analysis (D) and KEGG enrichment analysis (E) of DEmRNAs. (TIF 5392 KB)
Rights and permissions
About this article
Cite this article
Zhi, Y., Du, J., Qian, M. et al. Long non-coding RNA RP11-342L8.2, derived from RNA sequencing and validated via RT-qPCR, is upregulated and correlates with disease severity in psoriasis patients. Ir J Med Sci 191, 2643–2649 (2022). https://doi.org/10.1007/s11845-021-02882-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11845-021-02882-y