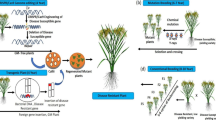
Abstract
Mildew Locus O (MLO), which encodes a seven-transmembrane domain protein, is responsible for the development of powdery mildew disease symptoms in many plant species. Hence, we knocked out PhMLO1 from Petunia hybrida cv. Mirage Rose at the genomic level using the Clustered regularly interspaced short palindromic repeat/CRISPR-associated nuclease 9 (CRISPR/Cas9) system and investigated involvement of the gene in powdery mildew disease development in the petunia. The MLO1 transcript levels observed in all mlo1 mutants were significantly lower than that in the wild type (WT). Specifically, the transcript level was the lowest in the mutants (line no. 1, 25, 33, and 85), followed by the mutants (line no. 14, 17, and 81), and line no. 6. Disease development was positively linked to the higher transcript levels of PhMLO1, because the disease severity observed in WT was greater than that observed in the mutants (line no. 6,14, 17, and 81), and disease symptoms were almost not observed in the mutants (line no. 1, 25, 33, and 85). In addition, the transmission of the edited alleles from the T0 to T1 generation was also confirmed, and the disease severity results were identical to those observed in the T0 mutant lines. Overall, the editing of PhMLO1 completely or partially prevented the development of this disease in this petunia, depending on MLO1 transcript level of the mutants. This study highlights the role of PhMLO1 in the development of powdery mildew disease in petunia and points to the need to edit the MLO genes in other ornamental plants to improve ornamental quality against powdery mildew disease.






Similar content being viewed by others
Availability of data and materials
Not applicable.
References
Ai TN, Naing AH, Arun M, Jeon SM, Kim CK (2017) Expression of RsMYB1 in petunia enhances anthocyanin production in vegetative and floral tissues. Sci Hortic 214:58–65
Bai Y, Pavan S, Zheng Z, Zappel NF, Reinstädler A, Lotti C, De Giovanni C, Ricciardi L, Lindhout P, Visser R (2008) Naturally occurring broad-spectrum powdery mildew resistance in a Central American tomato accession is caused by loss of Mlo function. Mol Plant Microbe Interact 21(1):30–39. https://doi.org/10.1094/MPMI-21-1-0030
Consonni C, Humphry ME, Hartmann HA, Livaja M, Durner J, Westphal L, Vogel J, Lipka V, Kemmerling B, Schulze-Lefert P (2006) Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet 38(6):716–720
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang D-L, Wang Z, Zhang Z, Zheng R, Yang L (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111(12):4632–4637
Frenkel O, Brewer MT, Milgroom MG (2010) Variation in pathogenicity and aggressiveness of Erysiphe necator from different Vitis spp. and geographic origins in the eastern United States. Phytopathology 100:1185–1193
Iovieno P, Andolfo G, Schiavulli A, Catalano D, Ricciardi L, Frusciante L, Ercolano MR, Pavan S (2015) Structure, evolution and functional inference on the Mildew Locus O (MLO) gene family in three cultivated Cucurbitaceae spp. BMC Genomics 16:1–13
Jiang P, Chen Y, Wilde HD (2016) Reduction of MLO1 expression in petunia increases resistance to powdery mildew. Sci Hortic 201:225–229
Jiwan D, Roalson EH, Main D, Dhingra A (2013) Antisense expression of peach mildew resistance locus O (PpMlo1) gene confers cross-species resistance to powdery mildew in Fragaria x ananassa. Transgenic Res 22:1119–1131
Jørgensen IH (1992) Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 63:141–152
Kaufmann H, Qiu X, Wehmeyer J, Debener T (2012) Isolation, molecular characterization, and mapping of four rose MLO orthologs. Front Plant Sci 3:244
Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS, Moon BC, Kang CH, Park CY, Yoo JH (2002) Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: Isolation and characterization of a rice Mlo homologue. J Biol Chem 277(22):19304–19314
Kim JM, Kim D, Kim S, Kim JS (2014) Genotyping with CRISPR-Casderived RNA-guided endonucleases. Nat Commun 5:3157
Naing AH, Park KI, Ai TN, Chung MY, Han JS, Kang YW, Lim KB, Kim CK (2017) Overexpression of snapdragon Delila (Del) gene in tobacco enhances anthocyanin accumulation and abiotic stress tolerance. BMC Plant Biol 17:1–4
Nekrasov V, Wang C, Win J, Lanz C, Weigel D, Kamoun S (2017) Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep 7(1):1–6
Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6(1):24765
Pépin N, Hebert FO, Joly DL (2021) Genome-Wide Characterization of the MLO Gene Family in Cannabis sativa Reveals Two Genes as Strong Candidates for Powdery Mildew Susceptibility. Front Plant Sci 12:729261
Pessina S, Pavan S, Catalano D, Gallotta A, Visser RG, Bai Y, Malnoy M, Schouten HJ (2014) Characterization of the MLO gene family in Rosaceae and gene expression analysis in Malus domestica. BMC Genomics 15:1–12
Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, Collins NC, Panstruga R, Schulze-Lefert P (2002) The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol 129(3):1076–1085
Punja ZK (2021) Emerging diseases of Cannabis sativa and sustainable management. Pest Manag Sci 77:3857–3870
Qiu X, Wang Q, Zhang H, Jian H, Zhou N, Ji C, Yan H, Bao M, Tang K (2015) Antisense RhMLO1 gene transformation enhances resistance to the powdery mildew pathogen in Rosa multiflora. Plant Mol Biol Report 33:1659–1665
Vielba-Fernández A, Polonio Á, Ruiz-Jiménez L, de Vicente A, Pérez-García A, Fernández-Ortuño D (2020) Fungicide resistance in powdery mildew fungi. Microorganisms 8(9):1431
Wan D-Y, Guo Y, Cheng Y, Hu Y, Xiao S, Wang Y, Wen Y-Q (2020) CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Horticulture research 7
Wang X, Tu M, Wang D, Liu J, Li Y, Li Z, Wang Y, Wang X (2018) CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol J 16(4):844–855
Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, Gao C, Qiu J-L (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32(9):947–951
Win KT, Zhang C, Lee S (2018) Genome-wide identification and description of MLO family genes in pumpkin (Cucurbita maxima Duch.). Hortic Environ Biotechnol 59:397–410
Xu J, Kang BC, Naing AH, Bae SJ, Kim JS, Kim H, Kim CK (2020) CRISPR/Cas9-mediated editing of 1-aminocyclopropane-1-carboxylate oxidase1 enhances Petunia flower longevity. Plant Biotechnol J 18(1):287–297
Xu J, Naing AH, Bunch H, Jeong J, Kim H, Kim CK (2021) Enhancement of the flower longevity of petunia by CRISPR/Cas9-mediated targeted editing of ethylene biosynthesis genes. Postharvest Biol Technol 174:111460
Yu J, Tu L, Subburaj S, Bae S, Lee G-J (2021) Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep 40:1037–1045
Zhang B, Xu X, Huang R, Yang S, Li M, Guo Y (2021) CRISPR/Cas9-mediated targeted mutation reveals a role for AN4 rather than DPL in regulating venation formation in the corolla tube of Petunia hybrida. Horticulture Research 8
Zhang B, Yang X, Yang C, Li M, Guo Y (2016) Exploiting the CRISPR/Cas9 system for targeted genome mutagenesis in petunia. Sci Rep 6(1):20315
Zheng Z, Nonomura T, Appiano M, Pavan S, Matsuda Y, Toyoda H, Wolters A-MA, Visser RG, Bai Y (2013) Loss of function in Mlo orthologs reduces susceptibility of pepper and tomato to powdery mildew disease caused by Leveillula taurica. PLoS ONE 8(7):e70723
Zhou SJ, Jing Z, Shi JL (2013) Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res 12:6565–6578
Acknowledgements
This work was carried out with the support of "Cooperative Research Program for Agriculture Science & Technology Development (Project No. RS-2022-RD010015)" Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Contributions
AHN and JX designed the study. WL constructed the vector and JX assisted the vector construction. MYC generated MLO1-edited transgenic lines. JX extracted DNA for Sanger sequencing and did powdery mildew test. JX produced T1 mutants and did powdery mildew test. AHN and JX analyzed the data. HK assisted the experiments. SYL helped fungal infection in the mutants. AHN wrote and revised the manuscript. CKK supervised the project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors report no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, J., Naing, A.H., Kang, H. et al. CRISPR/Cas9-mediated editing of PhMLO1 confers powdery mildew resistance in petunia. Plant Biotechnol Rep 17, 767–775 (2023). https://doi.org/10.1007/s11816-023-00854-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-023-00854-5