Abstract
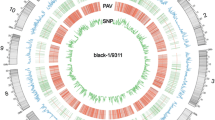
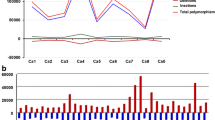
Citrus unshiu Marc. cv. Miyagawa-wase is the most widely cultivated citrus variety in Korea. To determine whether the C. unshiu genome used in this study shows genetic variation compared to the published reference genome (C. unshiu Marc. Miyagawa-wase), we conducted genome re-sequencing of two C. unshiu cultivars (Miyagawa-1 and Miyagawa-2) cultivated on Jeju in Korea. Compared with the reference genome, 1,198,650 and 1,207,084 single-nucleotide polymorphisms (SNPs) and 172,259 and 172,391 insertion/deletion polymorphisms (InDels) were detected in the Miyagawa-1 and -2 genomes, respectively. In SNP and InDel classifications by genome annotation, 367,591 and 369,068 SNPs and 45,362 and 45,464 InDels were located in the genic regions of Miyagawa-1 and -2, respectively. Among the SNPs of Miyagawa-1 and -2, transitions were more frequent than transversions. The majority of InDels was distributed in 1-bp InDels in both cultivars. The comparative number of total SNPs between Miyagawa-1 and -2 was smaller than the number of SNPs between the reference genome and Miyagawa-1 or -2. Gene ontology (GO) analysis showed that 23,164 and 23,049 genes with SNPs and 16,830 and 16,774 genes with InDels were annotated in the GO database. Taken together, Miyagawa-1 and -2 show genome-wide variation, including SNPs and InDels, compared to the published C. unshiu Marc. cv. Miyagawa-wase genome. This study suggests it would be more accurate to use the Miyagawa-1 and -2 genome sequences as a reference when conducting research using C. unshiu cultivated in Korea.




Similar content being viewed by others
References
Al Mamun AA, Lombardo MJ, Shee C, Lisewski AM, Gonzalez C, Lin D, Nehring RB, Saint-Ruf C, Gibson JL, Frisch RL, Lichtarge O, Hastings PJ, Rosenberg SM (2012) Identity and function of a large gene network underlying mutagenic repair of DNA breaks. Science 338:1344–1348
Aleza P, Juárez J, Hernández M, Pina JA, Ollitrault P, Navarro L (2009) Recovery and characterization of a Citrus clementina Hort. ex Tan. “Clemenules” haploid plant selected to establish the reference whole Citrus genome sequence. BMC Plant Biol 9:110
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H et al (2000) Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet 25(1):25–29
Baer CF, Miyamoto MM, Denver DR (2007) Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat Rev Genet 8:619–631
Baron E, Richirt J, Villoutreix R, Amsellem L, Roux F (2015) The genetics of intra- and interspecific competitive response and effect in a local population of an annual plant species. Funct Eco 29:1361–1370
Bartoli C, Roux F (2017) Genome-wide association studies in plant pathosystems: toward an ecological genomics approach. Front Plant Sci 8:763
Bohry D, Ramos HCC, Santos PHD, Boechat MSB, Arêdes FAS, Pirovani AAV, Pereira MG (2021) Discovery of SNPs and InDels in papaya genotypes and its potential for marker assisted selection of fruit quality traits. Sci Rep 11:292
Cox MP, Peterson DA, Biggs PJ (2010) SolexaQA: at-a-glance quality assessment of Illumina second-generation sequencing data. BMC Bioinform 11:485
De Kort H, Prunier JG, Ducatez S, Honnay O, Baguette M, Stevens VM, Blanchet S (2021) Life history, climate and biogeography interactively affect worldwide genetic diversity of plant and animal populations. Nat Commun 12:516
Du Y, Luo S, Li X, Yang J, Cui T, Li W, Lixia YL, Feng H, Chen Y, Mu J, Chen X, Shu Q, Guo T, Luo W, Zhou L (2017) Identifcation of substitutions and small insertion–deletions induced by carbon-ion beam irradiation in Arabidopsis thaliana. Front Plant Sci 8:1851
Ellstrand NC, Roose ML (1987) Patterns of genetic diversity in clonal plant species. Am J Bot 74(1):123–131
Healey A, Furtado A, Cooper T, Henry RJ (2014) Protocol: a simple method for extracting next-generation sequencing quality genomic DNA from recalcitrant plant species. Plant Methods 10:21
Jiang C, Mithani A, Belfield EJ, Mott R, Hurst LD, Harberd NP (2014) Environmentally responsive genome-wide accumulation of de novo Arabidopsis thaliana mutations and epimutations. Genome Res 24:1821–1829
Kim JE, Oh SK, Lee JH, Lee BM, Jo SH (2014) Genome-wide SNP calling using next generation sequencing data in tomato. Mol Cells 37:36–42
Kim SW, Ahn MS, Kwon YK, Song SY, Kim JK, Ha SH, Kim IJ, Liu JR (2015) Monthly metabolic changes and PLS prediction of carotenoid content of citrus fruit by combined Fourier transform infrared spectroscopy and quantitative HPLC analysis. Plant Biotechnol Rep 9:247–258
Kim WJ, Ryu J, Im J, Kim SH, Kang SY, Lee JH, Jo SH, Ha BK (2018) Molecular characterization of proton beam-induced mutations in soybean using genotyping-by-sequencing. Mol Genet Genomics 293:1169–1180
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J et al (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li Y, Cao K, Li N, Zhu G, Fang W, Chen C, Wang X, Guo J, Wang Q, Ding T, Wang J, Guan L, Wang J, Liu K, Guo W, Arús P, Huang S, Fei Z, Wang L (2021) Genomic analyses provide insights into peach local adaptation and responses to climate change. Genome Res 31:592–606
Liu TJ, Zhou JJ, Chen FY, Gan ZM, Li YP, Zhang JZ, Hu CG (2018) Identification of the genetic variation and gene exchange between Citrus Trifoliata and Citrus Clementina. Biomolecules 8:182
Martin M (2011) Cutadpat: cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J 17:10–12
Park HS, Sawamura M (2002) Comparison of the cold-pressed peel oil composition between Korean and Japanese Satsuma mandarin (Citrus unshiu Marcov. forma Miyagawa-wase) by GC, GC-MS and GC-O. J Food Sci Natr 7:5–11
Rando OJ, Verstrepen KJ (2007) Timescales of genetic and epigenetic inheritance. Cell 128:655–668
Sanczuk P, Govaert S, Meeussen C, De Pauw K, Vanneste T et al. (2021) Small scale environmental variation modulates plant defence syndromes of understorey plants in deciduous forests of Europe. Global Ecol Biogeogr 30:205–219
Shelake RM, Pramanik D, Kim JY (2019) Evolution of plant mutagenesis tools: a shifting paradigm from random to targeted genome editing. Plant Biotechnol Rep 13:423–445
Shimizu T, Tanizawa Y, Mochizuki T, Nagasaki H, Yoshioka T, Toyoda A, Fujiyama A, Kaminuma E, Nakamura Y (2017) Draft sequencing of the heterozygous diploid genome of satsuma (Citrus unshiu Marc.) using a hybrid assembly approach. Front Genet 8:180
Shinozaki K, Yamaguchi-Shinozaki K (1997) Gene expression and signal transduction in water-stress response. Plant Physiol 115:327–334
Shor E, Fox CA, Broach JR (2013) The yeast environmental stress response regulates mutagenesis induced by proteotoxic stress. PLoS Genet 9:e1003680
Subrahmaniam HJ, Libourel C, Journet EP, Morel JB, Munos S, Niebel A, Raffaele S, Roux F (2018) The genetics underlying natural variation of plant–plant interactions, a beloved but forgotten member of the family of biotic interactions. Plant J 93:747–770
Thudi M, Khan AW, Kumar V, Gaur PM, Katta K, Garg V, Roorkiwal M, Samineni S, Varshney RK (2016) Whole genome re-sequencing reveals genome-wide variations among parental lines of 16 mapping populations in chickpea (Cicer arietinum L.). BMC Plant Biol 16:10
Xu G, Chen L, Ruan X, Chen D, Zhu A et al (2012) The draft genome of sweet orange (Citrus sinensis). Nat Genet 45(1):59–67
Zhang QJ, Zhu T, Xia EH, Shi C, Liu YL, Zhang Y, Liu Y, Jiang WK, Zhao YJ, Mao SY et al (2014) Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc Natl Acad Sci USA 111:4954–4962
Acknowledgements
This research was supported by the 2021 Scientific Promotion Program funded by Jeju National University. The Subtropical Horticulture Research Institute and Research Institute for Subtropical Agriculture and Biotechnology provided facilities and equipment to conduct this research.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Eun, CH., Kim, IJ. Genome-wide DNA polymorphisms of Citrus unshiu Marc. cv. Miyagawa-wase cultivated in different regions based on whole-genome re-sequencing. Plant Biotechnol Rep 15, 551–559 (2021). https://doi.org/10.1007/s11816-021-00696-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-021-00696-z