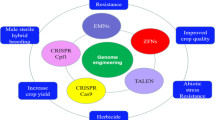
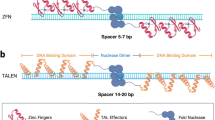
Abstract
Conventional breeding techniques for crop improvement are based on hybridization and selection. However, due to the long breeding cycles of crops and the potentially unpredictable effects of traditional breeding, these techniques are not sufficient to meet market demands for crops with a variety of traits or to address the emerging food crisis we could face in the near future. In the past decade, advanced technologies such as next-generation sequencing have been used to rapidly produce massive amounts of genome sequence information in many crop species. These techniques, together with targeted genome editing tools such as Zinc Finger Nuclease (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs), Clustered Regularly Interspaced Short Palindromic Sequences (CRISPR)/CRISPR-associated protein (Cas) have increased the possibilities for crop improvement via targeted genome editing. The use of these technologies in crop biology has opened up a new era of genome editing-mediated crop breeding. In this review, we summarize the current techniques used for site-directed genome editing in plants, focusing on the CRISPR/Cas system, and discuss their current and future applications for crop biology.





Similar content being viewed by others
References
Abdallah N, Prakash C, McHughen A (2015) Genome editing for crop improvement: Challenges and opportunities. GM Crops Food 6:183–205
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P (2007) CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712
Bhaya D, Davison M, Barrangou R (2011) CRISPR-Cas systems in bacteria and archaea: versatile small RNAs for adaptive defense and regulation. Annu Rev Genet 45:273–297
Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U (2009) Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326:1509–1512
Cai CQ, Doyon Y, Ainley WM, Miller JC, DeKelver RC, Moehle EA, Rock JM, Lee Y-L, Garrison R, Schulenberg L (2009) Targeted transgene integration in plant cells using designed zinc finger nucleases. Plant Mol Biol 69:699–709
Cai Y, Chen L, Liu X, Guo C, Sun S, Wu C, Jiang B, Han T, Hou W (2018) CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol J 16:176–185
Cermak T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJ, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL (2017) A multi-purpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini L (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Endo A, Masafumi M, Kaya H, Toki S (2016) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6:38169
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/C as-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359
Fineran PC, Charpentier E (2012) Memory of viral infections by CRISPR-Cas adaptive immune systems: acquisition of new information. Virology 434:202–209
Fonfara I, Richter H, Bratovič M, Le Rhun A, Charpentier E (2016) The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature 532:517–521
Gasiunas G, Barrangou R, Horvath P, Siksnys V (2012) Cas9–crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA 109:E2579–E2586
Gaudelli NM, Komor AC, Rees HA, Packer MS, Badran AH, Bryson DI, Liu DR (2017) Programmable base editing of A· T to G· C in genomic DNA without DNA cleavage. Nature 551:464–471
Hilscher J, Bürstmayr H, Stoger E (2017) Targeted modification of plant genomes for precision crop breeding. Biotechnol J 12:1600173
Hilton IB, D’ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, Gersbach CA (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol 33:510–517
Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ (2018) Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol J 16:1275–1282
Jaganathan D, Ramasamy K, Sellamuthu G, Jayabalan S, Venkataraman G (2018) CRISPR for crop improvement: an update review. Front Plant Sci 9:985
Jiang WZ, Henry IM, Lynagh PG, Comai L, Cahoon EB, Weeks DP (2017) Significant enhancement of fatty acid composition in seeds of the allohexaploid, Camelina sativa, using CRISPR/Cas9 gene editing. Plant Biotechnol J 15:648–657
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821
Khandagale K, Nadaf A (2016) Genome editing for targeted improvement of plants. Plant Biotechnol Rep 10:327–343
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR (2016) Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533:420–424
Li T, Liu B, Spalding MH, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392
Li J, Meng X, Zong Y, Chen K, Zhang H, Liu J, Li J, Gao C (2016) Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat Plant 2:16139
Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10:526–529
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018a) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:59
Li J, Zhang X, Sun Y, Zhang J, Du W, Guo X, Li S, Zhao Y, Xia L (2018b) Efficient allelic replacement in rice by gene editing: A case study of the NRT1.1B gene. J Integr Plant Biol 60:536–540
Li S, Li J, Zhang J, Du W, Fu J, Sutar S, Zhao Y, Xia L (2018c) Synthesis-dependent repair of Cpf1-induced double strand DNA breaks enables targeted gene replacement in rice. J Exp Bot 69:4715–4721
Lowder LG, Paul JW, Baltes NJ, Voytas DF, Zhang Y, Zhang D, Tang X, Zheng X, Hsieh T-F, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985
Lu Y, Zhu J-K (2017) Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system. Mol Plant 10:523–525
Lu K, Wu B, Wang J, Zhu W, Nie H, Qian J, Huang W, Fang Z (2018) Blocking amino acid transporter Os AAP 3 improves grain yield by promoting outgrowth buds and increasing tiller number in rice. Plant Biotechnol J 16:1710–1722
Lusser M, Parisi C, Plan D, Rodríguez-Cerezo E (2012) Deployment of new biotechnologies in plant breeding. Nat Biotechnol 30:231–239
Macovei A, Sevilla NR, Cantos C, Jonson GB, Slamet-Loedin I, Čermák T, Voytas DF, Choi IR, Chadha-Mohanty P (2018) Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol J 16:1918–1927
Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH (2015) An updated evolutionary classification of CRISPR–Cas systems. Nat Rev Microbiol 13:722–736
Maresca M, Lin VG, Guo N, Yang Y (2013) Obligate ligation-gated recombination (ObLiGaRe): custom-designed nuclease-mediated targeted integration through nonhomologous end joining. Genome Res 23:539–546
Miki D, Zhang W, Zeng W, Feng Z, Zhu J-K (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9:1967
Mishra R, Zhao K (2018) Genome editing technologies and their applications in crop Improvement. Plant Biotechnol Rep 12:57–68
Mladenov E, Iliakis G (2011) Induction and repair of DNA double strand breaks: the increasing spectrum of non-homologous end joining pathways. Mutat Res 711:61–72
Mojica FJ, Díez-Villaseñor C, García-Martínez J, Almendros C (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155:733–740
Nakayasu M, Akiyama R, Lee HJ, Osakabe K, Osakabe Y, Watanabe B, Sugimoto Y, Umemoto N, Saito K, Muranaka T (2018) Generation of α-solanine-free hairy roots of potato by CRISPR/Cas9 mediated genome editing of the St16DOX gene. Plant Physiol Biochem 131:70–77
Organisms EPoGM (2012) Scientific opinion addressing the safety assessment of plants developed using Zinc Finger Nuclease 3 and other Site-Directed Nucleases with similar function. EFSA J 10:2943
Ortigosa A, Gimenez-Ibanez S, Leonhardt N, Solano R (2018) Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J 1–9
Pabo CO, Peisach E, Grant RA (2001) Design and selection of novel Cys2His2 zinc finger proteins. Annu Rev Biochem 70:313–340
Peng A, Chen S, Lei T, Xu L, He Y, Wu L, Yao L, Zou X (2017) Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol J 15:1509–1519
Puchta H (2004) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14
Ray DK, Mueller ND, West PC, Foley JA (2013) Yield trends are insufficient to double global crop production by 2050. PLoS One 8:e66428
Ricroch A, Clairand P, Harwood W (2017) Use of CRISPR systems in plant genome editing: toward new opportunities in agriculture. Emerg Top Life Sci 1:169–182
Shah SA, Erdmann S, Mojica FJ, Garrett RA (2013) Protospacer recognition motifs: mixed identities and functional diversity. RNA Biol 10:891–899
Shan Q, Wang Y, Chen K, Liang Z, Li J, Zhang Y, Zhang K, Liu J, Voytas DF, Zheng X (2013) Rapid and efficient gene modification in rice and Brachypodium using TALENs. Mol Plant 6:1365–1368
Sharma S, Kaur R, Singh A (2017) Recent advances in CRISPR/Cas mediated genome editing for crop improvement. Plant Biotechnol Rep 11:193–207
Shimatani Z, Kashojiya S, Takayama M, Terada R, Arazoe T, Ishii H, Teramura H, Yamamoto T, Komatsu H, Miura K (2017) Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat Biotechnol 35:441
Shimatani Z, Fujikura U, Ishii H, Matsui Y, Suzuki M, Ueke Y, Taoka K-i, Terada R, Nishida K, Kondo A (2018) Inheritance of co-edited genes by CRISPR-based targeted nucleotide substitutions in rice. Plant Physiol Biochem 131:78–83
Silva G, Poirot L, Smith J, Montoya G, Duchateau P, Pâques F (2011) Meganucleases and other tools for targeted genome engineering: perspectives and challenges for gene therapy. Curr Gene Ther 11:11–27
Sprink T, Metje J, Hartung F (2015) Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr Opin Biotechnol 32:47–53
Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA (2014) DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507:62
Tang X, Zheng X, Qi Y, Zhang D, Cheng Y, Tang A, Voytas DF, Zhang Y (2016) A single transcript CRISPR-Cas9 system for efficient genome editing in plants. Mol Plant 9:1088–1091
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q (2017) A CRISPR–Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018
Tilman D, Balzer C, Hill J, Befort BL (2011) Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA 108:20260–20264
Van de Wiel C, Schaart J, Lotz L, Smulders M (2017) New traits in crops produced by genome editing techniques based on deletions. Plant Biotechnol Rep 11:1–8
Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64:327–350
Voytas DF, Joung JK (2009) DNA binding made easy. Science 326:1491–1492
Wang M, Mao Y, Lu Y, Tao X, Zhu J-k (2017) Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10:1011–1013
Waterworth WM, Drury GE, Bray CM, West CE (2011) Repairing breaks in the plant genome: the importance of keeping it together. New Phytol 192:805–822
Wright DA, Townsend JA, Winfrey RJ Jr., Irwin PA, Rajagopal J, Lonosky PM, Hall BD, Jondle MD, Voytas DF (2005) High-frequency homologous recombination in plants mediated by zinc-finger nucleases. Plant J 44:693–705
Xing H-L, Dong L, Wang Z-P, Zhang H-Y, Han C-Y, Liu B, Wang X-C, Chen Q-J (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol 14:327
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J (2017) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15:713–717
Yu Q-h, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q (2017) CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep 7:11874
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, Van Der Oost J, Regev A (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Zhang H, Zhang J, Lang Z, Botella JR, Zhu J-K (2017) Genome editing—principles and applications for functional genomics research and crop improvement. CRC Crit Rev Plant Sci 36:291–309
Zhao Y, Zhang C, Liu W, Gao W, Liu C, Song G, Li W-X, Mao L, Chen B, Xu Y (2016) An alternative strategy for targeted gene replacement in plants using a dual-sgRNA/Cas9 design. Sci Rep 6:23890
Zong Y, Wang Y, Li C, Zhang R, Chen K, Ran Y, Qiu J-L, Wang D, Gao C (2017) Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion. Nat Biotechnol 35:438–440
Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, Freschi L, Voytas DF, Kudla J, Peres LEP (2018) De novo domestication of wild tomato using genome editing. Nat Biotechnol. https://doi.org/10.1038/nbt.4272
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science and Technology Development (Project no. PJ01389401 to Y.J.)” Rural Development Administration, Republic of Korea and the National Research Foundation of Korea grant funded by the Korean Government (NRF-2016R1D1A1B03931167 to G.J.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jang, G., Joung, Y.H. CRISPR/Cas-mediated genome editing for crop improvement: current applications and future prospects. Plant Biotechnol Rep 13, 1–10 (2019). https://doi.org/10.1007/s11816-018-0509-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-018-0509-4