Abstract
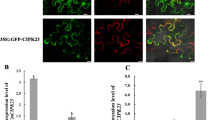
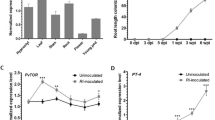
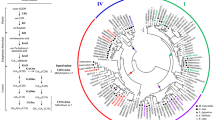
Rice is the principle staple food for more than half of humankind. Frequently, productivity of rice is affected by low nitrogen in the soil and hence, for enhanced rice production it heavily relies on synthetic nitrogen fertilizers that beget economic and ecological costs. In this context, the interest in transferring legume-like biological nitrogen fixation capability to rice has increased lately. The rice-arbuscular mycorrhizal (AM) symbiosis is mediated by genes that are orthologous to legume-genes known to be essential constituents of the common symbiotic pathway (CSP) that facilitates the establishment of both rhizobial nitrogen fixation- and AM-symbioses in legumes. Particularly, DMI3 (Ca+2/calmodulin-dependent serine/threonine protein kinase, CCaMK), a component of the CSP, was found to play a paramount role in promoting the development of both types of symbioses. In fact, expression of autoactive version of DMI3 was shown to be sufficient to trigger downstream developmental processes leading to the induction of spontaneous nodulation in the absence of rhizobia. Hence, in the present investigation, we expressed in transgenic rice a gain-of-function Medicago truncatula DMI3 T271D gene (gofMtDMI3) and assessed if legume-like symbiotic responses can be mimicked in rice roots. Ectopic expression of gofMtDMI3 in common bean induced spontaneous nodulation in the roots in the absence of rhizobia, but in rice plants it did not produce any such legume-like nodular manifestations. Conversely, the expression of gofMtDMI3 supported elevated AM colonization in rice roots that could improve plant nutrition/growth. In addition, gofMtDMI3 expression induced higher transcript levels of the CSP orthologues OsDMI3, OsIPD3 and OsNSP1, as well as triggered changes in the expression of several genes involved in biotic and abiotic stress responses. Our results with gofMtDMI3 lay the basis for the potential development of a biotechnological approach towards improvement of rice production.





Similar content being viewed by others
References
Akiyama K, Matsuzaki K, Hayashi H (2005) Plant sesquiterpenes induce hyphal branching in arbuscular mycorrhizal fungi. Nature 435:824–827. doi:10.1038/nature03608
Alexa A, Rahnenführer J (2016) topGO: enrichment analysis for gene ontology. R package version 2.28.0
Anders S, Huber W, Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M, Mortazavi A, Williams B, McCue K, Schaeffer L, Wold B, Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, Thiessen N, Griffith O, He A, Marra M, Snyder M, Jones S, Licatalosi D, Mele A, Fak J, Ule J, Kayikci M, Chi S, Clark T, Schweitzer A, Blume J, Wang X, Darnell J, Darnell R, Smith A, Heisler L, Mellor J, Kaper F, Thompson M, Chee M, Roth F, Giaever G, Nislow C, Marioni J, Mason C, Mane S, Stephens M, Gilad Y, Wang L, Feng Z, Wang X, Wang X, Zhang X, Robinson M, Smyth G, Whitaker L, Robinson M, McCarthy D, Smyth G, Robinson M, Smyth G, Cameron A, Trivedi P, Robinson M, Oshlack A, Loader C, McCullagh P, Nelder J, Agresti A, Engström P, Tommei D, Stricker S, Smith A, Pollard S, Bertone P, Morrissy A, Morin R, Delaney A, Zeng T, McDonald H, Jones S, Zhao Y, Hirst M, Marra M, Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak S, Habegger L, Rozowsky J, Shi M, Urban A, Hong M, Karczewski K, Huber W, Weissman S, Gerstein M, Korbel J, Snyder M, Benjamini Y, Hochberg Y, Bullard J, Purdom E, Hansen K, Dudoit S, Bloom J, Khan Z, Kruglyak L, Singh M, Caudy A, Smyth G, Smyth G, Lönnstedt I, Speed T, Gentleman R, Carey V, Bates D, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini A, Sawitzki G, Smith C, Smyth G, Tierney L, Yang J, Zhang J, Bliss C, Fisher R, Clark S, Perry J, Lawless J, Saha K, Paul S, Langmead B, Trapnell C, Pop M, Salzberg S (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. doi:10.1186/gb-2010-11-10-r106
Aparicio-Fabre R, Guillén G, Loredo M, Arellano J, Valdés-López O, Ramírez M, Iñiguez LP, Panzeri D, Castiglioni B, Cremonesi P, Strozzi F, Stella A, Girard L, Sparvoli F, Hernández G (2013) Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol 13:26. doi:10.1186/1471-2229-13-26
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G, Consortium GO (2000) Gene ontology: tool for the unification of biology. Nat Genet 25:25–29. doi:10.1038/75556
Banba M, Gutjahr C, Miyao A, Hirochika H, Paszkowski U, Kouchi H, Imaizumi-Anraku H (2008) Divergence of evolutionary ways among common sym genes: CASTOR and CCaMK show functional conservation between two symbiosis systems and constitute the root of a common signaling pathway. Plant Cell Physiol 49:1659–1671. doi:10.1093/pcp/pcn153
Beatty PH, Good AG (2011) Future prospects for Cereals. Science 333:416–418. doi:10.1126/science.1209467
Berruti A, Lumini E, Balestrini R, Bianciotto V (2016) Arbuscular mycorrhizal fungi as natural biofertilizers: let’s benefit from past successes. Front Microbiol 6:1–13. doi:10.3389/fmicb.2015.01559
Besserer A, Puech-Pagès V, Kiefer P, Gomez-Roldan V, Jauneau A, Roy S, Portais JC, Roux C, Bécard G, Séjalon-Delmas N (2006) Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol 4:1239–1247. doi:10.1371/journal.pbio.0040226
Bhattarai KK, Atamian HS, Kaloshian I, Eulgem T (2010) WRKY72-type transcription factors contribute to basal immunity in tomato and Arabidopsis as well as gene-for-gene resistance mediated by the tomato R gene Mi-1. Plant J 63:229–240. doi:10.1111/j.1365-313X.2010.04232.x
Boisson-Dernier A, Andriankaja A, Chabaud M, Niebel A, Journet EP, Barker DG, de Carvalho-Niebel F (2005) MtENOD11 gene activation during rhizobial infection and mycorrhizal arbuscule development requires a common AT-rich-containing regulatory sequence. Mol Plant Microbe Interact 18:1269–1276. doi:10.1094/MPMI-18-1269
Broughton WJ, Dilworth MJ (1971) Control of leghaemoglobin synthesis in snake beans. Biochem J 125:1075–1080. doi:10.1104/pp.112.197269
Burlat V, Kwon M, Davin LB, Lewis NG (2001) Dirigent proteins and dirigent sites in lignifying tissues. Phytochemistry 57:883–897. doi:10.1016/S0031-9422(01)00117-0
Chaintreuil C, Giraud E, Prin Y, Lorquin J, Bâ A, Gillis M, De Lajudie P, Chaintreuil M, Ba A (2000) Photosynthetic bradyrhizobia are natural endophytes of the African wild rice Oryza breviligulata. Appl Environ Microbiol 66:5437–5447. doi:10.1128/AEM.66.12.5437-5447.2000.Updated
Chen C, Gao M, Liu J, Zhu H (2007) Fungal symbiosis in rice requires an ortholog of a legume common symbiosis gene encoding a Ca2+/calmodulin-dependent protein kinase. Plant Physiol 145:1619–1628. doi:10.1104/pp.107.109876
Chen C, Ané JM, Zhu H (2008) OsIPD3, an ortholog of the Medicago truncatula DMI3 interacting protein IPD3, is required for mycorrhizal symbiosis in rice. New Phytol 180:311–315. doi:10.1111/j.1469-8137.2008.02612.x
Chen C, Fan C, Gao M, Zhu H (2009) Antiquity and function of CASTOR and POLLUX, the twin ion channel-encoding genes key to the evolution of root symbioses in plants. Plant Physiol 149:306–317. doi:10.1104/pp.108.131540
Chung S-M, Frankman EL, Tzfira T (2005) A versatile vector system for multiple gene expression in plants. Trends Plant Sci 10:357–361. doi:10.1016/j.tplants.2005.06.001
Dao TH, Linthorst H, Verpoorte R (2011) Chalcone synthase and its functions in plant resistance. Phytochem Rev 10:397–412. doi:10.1007/s11101-011-9211-7
de Vrieze J (2015) The littlest farmhands. Science 349:680–683. doi:10.1126/science.349.6249.680
Delaux PM, Radhakrishnan G, Oldroyd G (2015) Tracing the evolutionary path to nitrogen-fixing crops. Curr Opin Plant Biol 26:95–99. doi:10.1016/j.pbi.2015.06.003
Dellaporta S, Wood J, Hicks J (1983) A plant DNA minipreparation: version II. Plant Mol Biol Report 1:19–21
Denancé N, Szurek B, Noël LD (2014) Emerging functions of nodulin-like proteins in non-nodulating plant species. Plant Cell Physiol 55:469–474. doi:10.1093/pcp/pct198
Dénarié J, Debellé F, Promé J-C (1996) Rhizobium lipo-chitooligosaccharide nodulation factors: signaling molecules mediating recognition and morphogenesis. Annu Rev Biochem 65:503–535. doi:10.1146/annurev.bi.65.070196.002443
Estrada-Navarrete G, Alvarado-Affantranger X, Olivares J-E, Guillén G, Díaz-Camino C, Campos F, Quinto C, Gresshoff PM, Sanchez F (2007) Fast, efficient and reproducible genetic transformation of Phaseolus spp. by Agrobacterium rhizogenes. Nat Protoc 2:1819–1824. doi:10.1038/nprot.2007.259
Gelli M, Duo Y, Reddy Konda A, Zhang C, Holding D, Dweikat I (2014) Identification of differentially expressed genes between sorghum genotypes with contrasting nitrogen stress tolerance by genome-wide transcriptional profiling. BMC Genom 15:179. doi:10.1016/j.cbpc.2009.12.007
Giovannetti M, Mari A, Novero M, Bonfante P (2015) Early Lotus japonicus root transcriptomic responses to symbiotic and pathogenic fungal exudates. Front Plant Sci 6:480. doi:10.3389/fpls.2015.00480
Giraud E, Hannibal L, Fardoux J, Verméglio A, Dreyfus B (2000) Effect of Bradyrhizobium photosynthesis on stem nodulation of Aeschynomene sensitiva. Proc Natl Acad Sci USA 97:14795–14800. doi:10.1073/pnas.250484097
Giraud E, Moulin L, Vallenet D, Barbe V, Cytryn E, Avarre J-C, Jaubert M, Simon D, Cartieaux F, Prin Y, Bena G, Hannibal L, Fardoux J, Kojadinovic M, Vuillet L, Lajus A, Cruveiller S, Rouy Z, Mangenot S, Segurens B, Dossat C, Franck WL, Chang W-S, Saunders E, Bruce D, Richardson P, Normand P, Dreyfus B, Pignol D, Stacey G, Emerich D, Vermeglio A, Medigue C, Sadowsky M (2007) Legumes symbioses: absence of Nod genes in photosynthetic bradyrhizobia. Science 316:1307–1312. doi:10.1126/science.1139548
Gleason C, Chaudhuri S, Yang TB, Munoz A, Poovaiah BW, Oldroyd GED (2006) Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441:1149–1152. doi:10.1038/Nature04812
Godfroy O, Debellé F, Timmers T, Rosenberg C (2006) A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol Plant Microbe Interact 19:495–501. doi:10.1094/MPMI-19-0495
Gomez-Roldan V, Fermas S, Brewer PB, Puech-Pagès V, Dun EA, Pillot J-P, Letisse F, Matusova R, Danoun S, Portais J-C, Bouwmeester H, Bécard G, Beveridge CA, Rameau C, Rochange SF (2008) Strigolactone inhibition of shoot branching. Nature 455:189–194. doi:10.1038/nature07271
Groten K, Nawaz A, Nguyen NHT, Santhanam R, Baldwin IT (2015) Silencing a key gene of the common symbiosis pathway in Nicotiana attenuata specifically impairs arbuscular mycorrhizal infection without influencing the root-associated microbiome or plant growth. Plant Cell Environ 38:2398–2416. doi:10.1111/pce.12561
Gutiérrez RA (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336:1673–1675. doi:10.1126/science.1217620
Gutjahr C, Banba M, Croset V, An K, Miyao A, An G, Hirochika H, Imaizumi-Anraku H, Paszkowski U (2008) Arbuscular mycorrhiza-specific signaling in rice transcends the common symbiosis signaling pathway. Plant Cell 20:2989–3005. doi:10.1105/tpc.108.062414
Gutjahr C, Radovanovic D, Geoffroy J, Zhang Q, Siegler H, Chiapello M, Casieri L, An K, An G, Guiderdoni E, Kumar CS, Sundaresan V, Harrison MJ, Paszkowski U (2012) The half-size ABC transporters STR1 and STR2 are indispensable for mycorrhizal arbuscule formation in rice. Plant J 69:906–920. doi:10.1111/j.1365-313X.2011.04842.x
Hakeem KR, Chandna R, Ahmad A, Qureshi MI, Iqbal M (2012) Proteomic analysis for low and high nitrogen-responsive proteins in the leaves of rice genotypes grown at three nitrogen levels. Appl Biochem Biotechnol 168:834–850. doi:10.1007/s12010-012-9823-4
Hayashi T, Banba M, Shimoda Y, Kouchi H, Hayashi M, Imaizumi-Anraku H (2010) A dominant function of CCaMK in intracellular accommodation of bacterial and fungal endosymbionts. Plant J 63:141–154. doi:10.1111/j.1365-313X.2010.04228.x
Hirsch S, Kim J, Muñoz A, Heckmann AB, Downie JA, Oldroyd GED (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21:545–557. doi:10.1105/tpc.108.064501
Ivleva NB, Groat J, Staub JM, Stephens M (2016) Expression of active subunit of nitrogenase via integration into plant organelle genome. PLoS One 11:1–13. doi:10.1371/journal.pone.0160951
Jach G, Binot E, Frings S, Luxa K, Schell J (2001) Use of red fluorescent protein from Discosoma sp. (dsRED) as a reporter for plant gene expression. Plant J 28:483–491. doi:10.1046/j.1365-313X.2001.01153.x
Jang I-C, Choi W-B, Lee K-H, Song SI, Nahm BH, Kim J-K (2002) High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol 129:1473–1481. doi:10.1104/pp.002261.1
Jefferson RA, Burgess SM, Hirsh D (1986) Beta-Glucuronidase from Escherichia coli as a gene-fusion marker. Proc Natl Acad Sci USA 83:8447–8451. doi:10.1073/pnas.83.22.8447
Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6:4. doi:10.1186/1939-8433-6-4
Ladha J, Reddy P (1995) Extension of nitrogen fixation to rice—necessity and possibilities. GeoJournal 35:363–372
Ladha J, Reddy P (2003) Nitrogen fixation in rice systems: state of knowledge and future prospects. Plant Soil 252:151–167. doi:10.1023/A:1024175307238
Langmead B, Trapnell C, Pop M, Salzberg S (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. doi:10.1186/gb-2009-10-3-r25
Lauressergues D, Delaux PM, Formey D, Lelandais-Brière C, Fort S, Cottaz S, Bécard G, Niebel A, Roux C, Combier JP (2012) The microRNA miR171 h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J 72:512–522. doi:10.1111/j.1365-313X.2012.05099.x
Lévy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet E-P, Ané J-M, Lauber E, Bisseling T, Dénarié J, Rosenberg C, Debellé F (2004) A putative Ca+2 and calmodulin- dependent protein kinase required. Science 303:1361–1364. doi:10.1126/science.1093038
Liu X, Bai X, Wang X, Chu C (2007) OsWRKY71, a rice transcription factor, is involved in rice defense response. J Plant Physiol 164:969–979. doi:10.1016/j.jplph.2006.07.006
Liu W, Kohlen W, Lillo A, Op den Camp R, Ivanov S, Hartog M, Limpens E, Jamil M, Smaczniak C, Kaufmann K, Yang W-C, Hooiveld GJEJ, Charnikhova T, Bouwmeester HJ, Bisseling T, Geurts R (2011) Strigolactone biosynthesis in Medicago truncatula and rice requires the symbiotic GRAS-type transcription factors NSP1 and NSP2. Plant Cell 23:3853–3865. doi:10.1105/tpc.111.089771
López-Torrejón G, Jiménez-Vicente E, Buesa JM, Hernandez JA, Verma HK, Rubio LM (2016) Expression of a functional oxygen-labile nitrogenase component in the mitochondrial matrix of aerobically grown yeast. Nat Commun 7:11426. doi:10.1038/ncomms11426
Madsen LH, Tirichine L, Jurkiewicz A, Sullivan JT, Heckmann AB, Bek AS, Ronson CW, James EK, Stougaard J (2010) The molecular network governing nodule organogenesis and infection in the model legume Lotus japonicus. Nat Commun 1:10. doi:10.1038/ncomms1009
Maillet F, Poinsot V, André O, Puech-Pagès V, Haouy A, Gueunier M, Cromer L, Giraudet D, Formey D, Niebel A, Martinez EA, Driguez H, Bécard G, Dénarié J (2011) Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature 469:58–63. doi:10.1038/nature09622
Markmann K, Parniske M (2009) Evolution of root endosymbiosis with bacteria: how novel are nodules? Trends Plant Sci 14:77–86. doi:10.1016/j.tplants.2008.11.009
Markmann K, Giczey G, Parniske M (2008) Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol 6:0497–0506. doi:10.1371/journal.pbio.0060068
McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA (1990) A new method which gives an objective measure of colonization of roots by vesicular- arbuscular mycorrhizal fungi. New Phytol 115:495–501. doi:10.1111/j.1469-8137.1990.tb00476.x
Molouba F, Lorquin J, Willems A, Hoste B, Giraud E, Dreyfus B, Gillis M, De Lajudie P, Masson-Boivin C (1999) Photosynthetic bradyrhizobia from Aeschynomene spp. are specific to stem-nodulated species and form a separate 16S ribosomal DNA restriction fragment length polymorphism group. Appl Environ Microbiol 65:3084–3094
Mornico D, Miché L, Béna G, Nouwen N, Verméglio A, Vallenet D, Smith AAT, Giraud E, Médigue C, Moulin L (2012) Comparative genomics of Aeschynomene symbionts: insights into the ecological lifestyle of nod-independent photosynthetic bradyrhizobia. Genes 3:35–61. doi:10.3390/genes3010035
Mus F, Crook MB, Garcia K, Garcia Costas A, Geddes BA, Kouri ED, Paramasivan P, Ryu M-H, Oldroyd GED, Poole PS, Udvardi MK, Voigt CA, Ané J-M, Peters JW (2016) Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol 82:3698–3710. doi:10.1128/AEM.01055-16.Editor
Narsai R, Ivanova A, Ng S, Whelan J (2010) Defining reference genes in Oryza sativa using organ, development, biotic and abiotic transcriptome datasets. BMC Plant Biol 10:56. doi:10.1186/1471-2229-10-56
Ohta S, Mita S, Hattori T, Nakamura K (1990) Construction and expression in tobacco of a {beta}-glucuronidase (GUS) reporter gene containing an intron within the coding sequence. Plant Cell Physiol 31:805–813
Oldroyd GED (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252–263. doi:10.1038/nrmicro2990
Petti C, Khan M, Doohan F (2010) Lipid transfer proteins and protease inhibitors as key factors in the priming of barley responses to Fusarium head blight disease by a biocontrol strain of Pseudomonas fluorescens. Funct Integr Genom 10:619–627. doi:10.1007/s10142-010-0177-0
Pimprikar P, Carbonnel S, Paries M, Katzer K, Klingl V, Bohmer MJ, Karl L, Floss DS, Harrison MJ, Parniske M, Gutjahr C (2016) A CCaMK-CYCLOPS-DELLA complex activates transcription of RAM1 to regulate arbuscule branching. Curr Biol 26:987–998. doi:10.1016/j.cub.2016.01.069
Reddy P, James E, Ladha J (2002) Nitrogen Fixation in Rice. In: Leigh GJ (ed) Nitrogen fixation at the millenium. Elsevier, Amsterdam, pp 421–445
Reddy P, Rendón-Anaya M, Soto del Río M, Khandual S (2007) Flavonoids as signaling molecules and regulators of root nodule development. Dyn Soil Dyn Plant 1(2):83–94
Reddy PM, Altúzar-Molina AR, Ortiz-Berrocal M, Medina-Andrés R, López-Sámano M, Martínez L (2013) Predisposition and redesigning of genetic networks of rice for accommodating nitrogen-fixing rhizobial symbiosis. In: Muralidharan K, Siddiq EA (eds) International dialogue on perception and prospects of designer rice. Society for Advancement of Rice Research, India, pp 245–257
Rogers C, Oldroyd GED (2014) Synthetic biology approaches to engineering the nitrogen symbiosis in cereals. J Exp Bot 65:1939–1946. doi:10.1093/jxb/eru098
Rose CM, Venkateshwaran M, Volkening JD, Grimsrud PA, Maeda J, Bailey DJ, Park K, Howes-Podoll M, den Os D, Yeun LH, Westphall MS, Sussman MR, Ané J-M, Coon JJ (2012) Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol Cell Proteom 11:724–744. doi:10.1074/mcp.M112.019208
Samac DA, Graham MA (2007) Recent advances in legume-microbe interactions: recognition, defense response, and symbiosis from a genomic perspective. Plant Physiol 144:582–587. doi:10.1104/pp.107.096503
Siciliano V, Genre A, Balestrini R, Cappellazzo G, deWit PJGM, Bonfante P (2007) Transcriptome analysis of arbuscular mycorrhizal roots during development of the prepenetration apparatus. Plant Physiol 144:1455–1466. doi:10.1104/pp.107.097980
Singh S, Parniske M (2012) Activation of calcium- and calmodulin-dependent protein kinase (CCaMK), the central regulator of plant root endosymbiosis. Curr Opin Plant Biol 15:444–453. doi:10.1016/j.pbi.2012.04.002
Sreevidya VS, Hernandez-Oane RJ, So RB, Sullia SB, Stacey G, Ladha JK, Reddy PM (2005) Expression of the legume symbiotic lectin genes psl and gs52 promotes rhizobial colonization of roots in rice. Plant Sci 169:726–736. doi:10.1016/j.plantsci.2005.05.024
Sreevidya VS, Srinivasa Rao C, Sullia SB, Ladha JK, Reddy PM (2006) Metabolic engineering of rice with soybean isoflavone synthase for promoting nodulation gene expression in rhizobia. J Exp Bot 57:1957–1969. doi:10.1093/jxb/erj143
Stokstad E (2016) The nitrogen fix. Science 353:1225–1227. doi:10.1126/science.353.6305.1225
Takeda N, Maekawa T, Hayashi M (2012) Nuclear-localized and deregulated calcium- and calmodulin-dependent protein kinase activates rhizobial and mycorrhizal responses in Lotus japonicus. Plant Cell 24:810–822. doi:10.1105/tpc.111.091827
Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M (2004) MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J 37:914–939. doi:10.1111/j.1365-313X.2004.02016.x
Tholl D (2006) Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol 9:297–304. doi:10.1016/j.pbi.2006.03.014
Tirichine L, James EK, Sandal N, Stougaard J (2006) Spontaneous root-nodule formation in the model legume Lotus japonicus: a novel class of mutants nodulates in the absence of rhizobia. Mol Plant-Microbe Interact 19:373–382. doi:10.1094/MPMI-19-0373
Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H (2006) Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J 47:969–976. doi:10.1111/j.1365-313X.2006.02836.x
Van Damme EJM, Barre A, Rougé P, Peumans WJ (2004) Cytoplasmic/nuclear plant lectins: a new story. Trends Plant Sci 9:484–489. doi:10.1016/j.tplants.2004.08.003
Venkateshwaran M, Volkening JD, Sussman MR, Ané J-M (2013) Symbiosis and the social network of higher plants. Curr Opin Plant Biol 16:118–127. doi:10.1016/j.pbi.2012.11.007
Weidenbach D, Esch L, Möller C, Hensel G, Kumlehn J, Höfle C, Hückelhoven R, Schaffrath U (2016) Polarized defense against fungal pathogens is mediated by the jacalin-related lectin domain of modular poaceae-specific proteins. Mol Plant 9:514–527. doi:10.1016/j.molp.2015.12.009
Werner GDA, Cornwell WK, Sprent JI, Kattge J, Kiers ET (2014) A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat Commun 5:1–9. doi:10.1038/ncomms5087
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493. doi:10.1104/pp.126.2.485
Xie Z, Zhang ZL, Xiaolu Z, Huang J, Ruas P, Thompson D, Shen QJ (2005) Annotations and functional analyses of the rice WRKY gene superfamily reveal positive and negative regulators of abscisic acid signaling in aleurone cells. Plant Physiol 137:176–189. doi:10.1104/pp.104.054312
Yokota K, Soyano T, Kouchi H, Hayashi M (2010) Function of GRAS proteins in root nodule symbiosis is retained in homologs of a non-legume, rice. Plant Cell Physiol 51:1436–1442. doi:10.1093/pcp/pcq124
Yoshida S, Forno DA, Cock JH, Gomez KA (1976) Laboratory manual for physiological Studies of Rice. International Rice Research Institute, Los Baños, pp 61–64
Yuan M, Wang S (2013) Rice MtN3/saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant 6:665–674. doi:10.1093/mp/sst035
Acknowledgements
M Ortiz-Berrocal is a doctoral student from Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and received a fellowship (No. 390781) from CONACYT. This research was supported by the grants from DGAPA (IN206208) and CONACyT (128135) to PM Reddy. We gratefully acknowledge Dr. Tzvi Tzfira (University of Michigan, Ann Arbor, USA) for the kind gift of SAT and RCS2-HPT vector systems and QFB Lourdes Martínez Aguilar (Centro de Ciencias Genómicas-UNAM) for her technical assistance.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ortiz-Berrocal, M., Lozano, L., Sanchez-Flores, A. et al. Expression in rice of an autoactive variant of Medicago truncatula DMI3, the Ca+2/calmodulin-dependent protein kinase from the common symbiotic pathway modifies root transcriptome and improves mycorrhizal colonization. Plant Biotechnol Rep 11, 271–287 (2017). https://doi.org/10.1007/s11816-017-0449-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-017-0449-4