Abstract
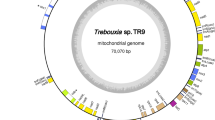
We determined the complete mitochondrial genome (mtDNA, mitogenome) of Pyropia tenera (Bangiales, Rhodophyta). Pyropia tenera mtDNA had a larger size (42,268 bp) than the mtDNA sequences of Porphyra and Pyropia reported previously, and encoded two ribosomal RNA genes [large subunit (rnl), small subunit (rns)], 24 transfer RNAs, four ribosomal proteins, and 17 genes involved in electron transport and oxidative phosphorylation. Moreover, four conserved open reading frames (ORFs) and six intronic ORFs (three in rnl and three in the cox1 gene) were also identified. In comparison with other Porphyra and Pyropia species, Py. tenera had four major structural changes in two gene loss/rearrangement regions [tRNA-Gln(uug)–tRNA-SeC(uca) and tRNA-Ala(ugc)–tRNA-Arg(ucu)] and two different patterns of exon, intron, and intronic ORFs (rnl and cox1). The unique features of Py. tenera mtDNA include the complete sequence of red algal mtDNA for investigating mtDNA evolution and developing molecular markers for species identification. In addition, red algal mtDNA can provide useful genetic information as a genetic reservoir for bioengineering.



Similar content being viewed by others
References
Bullerwell CE, Gray MW (2004) Evolution of the mitochondrial genome: protist connections to animals, fungi and plants. Curr Opin Microbiol 7:528–534
Burger G, Saint-Louis D, Gray MW, Lang BF (1999) Complete sequence of the mitochondrial DNA of the red alga Porphyra purpurea: Cyanobacterial introns and shared ancestry of red and green algae. Plant Cell 11:1675–1694
Conant GC, Wolfe KH (2008) GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics 24:861–862
Dai L, Toor N, Olson R, Keeping A, Zimmerly S (2003) Database for mobile group II introns. Nucleic Acids Res 31:424–426
Hancock L, Goff L, Lane C (2010) Red algae lose key mitochondrial genes in response to becoming parasitic. Genome Biol Evol 2:897–910
Hwang MS, Kim S-M, Ha D-S, Baek JM, Kim H-S, Choi H-G (2005) DNA sequences and identification of Porphyra cultivated by natural seeding on the southwest coast of Korea. Algae 20:183–196
Ikuta K, Kawai H, Müller DG, Ohama T (2008) Recurrent invasion of mitochondrial group II Introns in specimens of Pylaiella littoralis (Brown Alga), collected worldwide. Curr Genet 53:207–216
Leblanc C, Boyen C, Richard O, Bonnard G, Grienenberger JM, Kloareg B (1995) Complete sequence of the mitochondrial DNA of the rhodophyte Chondrus crispus (Gigartinales). Gene content and genome organization. J Mol Biol 250:484–495
Lin C-P, Wu C-S, Huang Y-Y, Chaw S-M (2012) The complete chloroplast genome of Ginkgo biloba reveals the mechanism of inverted repeat contraction. Genome Biol Evol 4:374–381
Mao Y, Zhang B, Kong F, Wang L (2012) The complete mitochondrial genome of Pyropia haitanensis Chang et Zheng. Mitochondrial DNA 23:344–346
Niwa K, Sakamoto T (2010) Allopolyploidy in natural and cultivated populations of Porphyra (Bangiales, Rhodophyta). J Phycol 46:1097–1105
Niwa K, Kobiyama A, Aruga Y (2005) Confirmation of cultivated Porphyra tenera (Bangiales, Rhodophyta) by polymerase chain reaction restriction fragment length polymorphism analyses of the plastid and nuclear DNA. Phycol Res 53:296–302
Odintsova MS, Yurina NP (2002) The mitochondrial genome of protists. Russ J Genet 38:642–655
Odom OW, Herrin DL (2010) An unprecedented group-II intron from Chlamydomonas caudata (Chlorophyceae) that has two large open reading frames-potentially encoding a reverse transcriptase-maturase-endonuclease and a novel protein. J Phycol 46:969–973
Ohta N, Sato N, Kuroiwa T (1998) Structure and organization of the mitochondrial genome of the unicellular red alga Cyanidioschyzon merolae deduced from the complete nucleotide sequence. Nucleic Acids Res 26:5190–5198
Park E-J, Fukuda S, Endo H, Kitade Y, Saga N (2007) Genetic polymorphism within Porphyra yezoensis (Bangiales, Rhodophyta) and related species from Japan and Korea detected by cleaved amplified polymorphic sequence analysis. Eur J Phycol 42:29–40
Sacerdot C, Fayat G, Dessen P, Springer M, Plumbridge JA, Grunberg-Manago M, Blanquet S (1982) Sequence of a 1.26-kb DNA fragment containing the structural gene for E. coli initiation factor IF3: presence of an AUU initiator codon. EMBO J 1:311–315
Schattner P, Brooks AN, Lowe TM (2005) The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res 33:W686–W689
Sellem CH, d’Aubenton-Carafa Y, Rossignol M, Belcour L (1996) Mitochondrial intronic open reading frames in Podospora: mobility and consecutive exonic sequence variations. Genetics 143:777–788
Shaw J, Lickey EB, Beck JT et al (2005) The tortoise and the hare II: relative utility of 21 noncoding chloroplast DNA sequences for phylogenetic analysis. Am J Bot 92:142–166
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Smith DR, Hua J, Lee RW, Keeling PJ (2012) Relative rates of evolution among the three genetic compartments of the red alga Porphyra differ from those of green plants and do not correlate with genome architecture. Mol Phylogenet Evol 65:33–344
Sutherland JE et al (2011) A new look at an ancient order: generic revision of the Bangiales (Rhodophyta). J Phycol 47:1131–1151
Swofford DL (2001) PAUP*. Phylogenetic Analysis using Parsimony (*and Other Methods). Version 4. Sinauer, Sunderland
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Wolf PG, Rowe CA, Sinclair RB, Hasebe M (2003) Complete nucleotide sequence of the chloroplast genome from a leptosporangiate fern, Adiantum capillus-veneris L. DNA Res 10:59–65
Xu X, Wu X, Yu Z (2012) Comparative studies of the complete mitochondrial genomes of four Paphia clams and reconsideration of subgenus Neotapes (Bivalvia: Veneridae). Gene 494:17–23
Acknowledgments
This study was supported by a grant (RP-2013-BT-031) from the National Fisheries Research and Development Institute, Korea.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hwang, M.S., Kim, SO., Ha, DS. et al. Complete sequence and genetic features of the mitochondrial genome of Pyropia tenera (Rhodophyta). Plant Biotechnol Rep 7, 435–443 (2013). https://doi.org/10.1007/s11816-013-0281-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11816-013-0281-4