Abstract
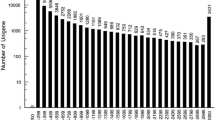
Chiton (Ischnochiton hakodadensis) is one of marine mollusks well known for its eight separate shell plates. I. hakodadensis is important, which plays a vital role in the ecosystems it inhabits. So far, the genetic studies on the chiton are scarce due in part to insufficient genomic resources available for this species. In this study, we investigated the transcriptome of the chiton foot using Illumina sequencing technology. The reads were assembled and clustered into 256461 unigenes, of which 42247 were divided into diverse functional categories by Gene Ontology (GO) annotation terms, and 17256 mapped onto 365 pathways by KEGG pathway mapping. Meanwhile, a set of differentially expressed genes (DEGs) between distal and proximal muscles were identified as the foot adhesive locomotion associated, thus were useful for our future studies. Moreover, up to 679384 high-quality single nucleotide polymorphisms (SNPs) and 19814 simple sequence repeats (SSRs) were identified in this study, which are valuable for subsequent studies on genetic diversity and variation. The transcriptomic resource obtained in this study should aid to future genetic and genomic studies of chiton.
Similar content being viewed by others
References
Beeley, J. G., 1985. Glycoprotein and Proteoglycan Techniques. Elsevier, Amsterdam, 6pp.
Benjamini, Y., and Hochberg, Y., 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. Journal of The Royal Statistical Society. Series B (Methodological), 57 (1): 289–300.
Bettencourt, R., Pinheiro, M., Egas, C., Gomes, P., Afonso, M., Shank, T., and Santos, R. S., 2010. High-throughput sequencing and analysis of the gill tissue transcriptome from the deep-sea hydrothermal vent mussel Bathymodiolus azoricus. BMC Genomics, 11: 559.
Byern, D. B. J. V., and Grunwald, D. B. I., 2010. Biological Adhesive Systems: From Nature to Technical and Medical Application. Springer, Vienna, 53pp.
Connors, M. J., Ehrlich, H., Hog, M., Godeffroy, C., Araya, S., Kallai, I., Gazit, D., Boyce, M., and Ortiz, C., 2012. Threedimensional structure of the shell plate assembly of the chiton Tonicella marmorea and its biomechanical consequences. Journal of Structural Biology, 177 (2): 314–328.
Craft, J. A., Gilbert, J. A., Temperton, B., Dempsey, K. E., Ashelford, K., Tiwari, B., Hutchinson, T. H., and Chipman, J. K., 2010. Pyrosequencing of Mytilus galloprovincialis cDNAs: Tissue-specific expression patterns. PLoS One, 5 (1): e8875.
D’Arrigo, C., Burl, S., Withers, A. P., Dobson, H., Black, C., and Boxer, M., 1998. TGF-beta1 binding protein-like modules of fibrillin-1 and -2 mediate integrin-dependent cell adhesion. Connective Tissue Research, 37 (1-2): 29–51.
Du, H., Bao, Z., Hou, R., Wang, S., Su, H., Yan, J., Tian, M., Li, Y., Wei, W., and Lu, W., 2012. Transcriptome sequencing and characterization for the sea cucumber Apostichopus japonicus (Selenka, 1867). PLoS One, 7 (3): e33311.
Fu, X., Sun, Y., Wang, J., Xing, Q., Zou, J., Li, R., Wang, Z., Wang, S., Hu, X., and Zhang, L., 2014. Sequencing-based gene network analysis provides a core set of gene resource for understanding thermal adaptation in Zhikong scallop Chlamys farreri. Molecular Ecology Resources, 14 (1): 184–198.
Götz, S., García-Gómez, J. M., Terol, J., Williams, T. D., Nagaraj, S. H., Nueda, M. J., Robles, M., Talón, M., Dopazo, J., and Conesa, A., 2008. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Research, 36 (10): 3420–3435.
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., Adiconis, X., Fan, L., Raychowdhury, R., and Zeng, Q., 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nature biotechnology, 29 (7): 644–652.
Hou, R., Bao, Z., Wang, S., Su, H., Li, Y., Du, H., Hu, J., Wang, S., and Hu, X., 2011. Transcriptome sequencing and de novo analysis for Yesso scallop (Patinopecten yessoensis) using 454 GS FLX. PLoS One, 6 (6): e21560.
Hu, X., Bao, Z., Hu, J., Shao, M., Zhang, L., Bi, K., Zhan, A., and Huang, X., 2006. Cloning and characterization of tryptophan 2, 3-dioxygenase gene of Zhikong scallop Chlamys farreri (Jones and Preston 1904). Aquaculture Research, 37 (12): 1187–1194.
Huan, P., Wang, H., and Liu, B., 2012. Transcriptomic analysis of the clam Meretrix meretrix on different larval stages. Marine Biotechnology, 14 (1): 69–78.
Johnstone, I. L., 1994. The cuticle of the nematode Caenorhabditis elegans: A complex collagen structure. Bioessays, 16 (3): 171–178.
Joubert, C., Piquemal, D., Marie, B., Manchon, L., Pierrat, F., Zanellacléon, I., Cochenneclaureau, N., Gueguen, Y., and Montagnani, C., 2010. Transcriptome and proteome analysis of Pinctada margaritifera calcifying mantle and shell: Focus on biomineralization. BMC Genomics, 11: 613.
Kangas, M., and Shepherd, S. A., 2013. Distribution and feeding of chitons in a boulder habitat at West Island, South Australia. Journal of the Malacological Society of Australia, 6 (3): 101–111.
Kielty, C. M., Baldock, C., Lee, D., Rock, M. J., Ashworth, J. L., and Shuttleworth, C. A., 2002. Fibrillin: From microfibril assembly to biomechanical function. Philosophical Transactions of the Royal Society B Biological Sciences, 357 (1418): 207–217.
Kielty, C. M., Wess, T. J., Haston, L., Ashworth, J. L., Sherratt, M. J., and Shuttleworth, C. A., 2003. Fibrillin-rich Microfibrils: Elastic Biopolymers of the Extracellular Matrix. Springer, Netherlands, 581–596.
Kinoshita, S., Wang, N., Inoue, H., Maeyama, K., Okamoto, K., Nagai, K., Kondo, H., Hirono, I., Asakawa, S., and Watabe, S., 2011. Deep sequencing of ESTs from nacreous and prismatic layer producing tissues and a screen for novel shell formation-related genes in the pearl oyster. PLoS One, 6 (6): e21238.
Koboldt, D. C., Zhang, Q., Larson, D. E., Dong, S., Mclellan, M. D., Ling, L., Miller, C. A., Mardis, E. R., Li, D., and Wilson, R. K., 2012. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research, 22 (3): 568–576.
Li, H., Ding, G. L., and Li, Y., 2009. PAnnBuilder: An R package for assembling proteomic annotation data. Bioinformatics, 25 (8): 1094–1095.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., and Durbin, R., 2009. The sequence alignment/map format and SAMtools. Bioinformatics, 25 (16): 2078–2079.
Liu, S., Zhou, Z., Lu, J., Sun, F., Wang, S., Hong, L., Jiang, Y., Kucuktas, H., Kaltenboeck, L., and Peatman, E., 2011. Generation of genome-scale gene-associated SNPs in catfish for the construction of a high-density SNP array. BMC Genomics, 12: 53.
Miao, Y., Zhang, L., Sun, Y., Jiao, W., Li, Y., Sun, J., Wang, Y., Wang, S., Bao, Z., and Liu, W., 2015. Integration of transcriptomic and proteomic approaches provides a core set of genes for understanding of scallop attachment. Marine Biotechnology, 17 (5): 523–532.
Milan, M., Coppe, A., Reinhardt, R., Cancela, L. M., Leite, R. B., Saavedra, C., Ciofi, C., Chelazzi, G., Patarnello, T., and Bortoluzzi, S., 2011. Transcriptome sequencing and microarray development for the Manila clam, Ruditapes philippinarum: Genomic tools for environmental monitoring. BMC Genomics, 12: 234.
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M., 2007. KAAS: An automatic genome annotation and pathway reconstruction server. Nucleic Acids Research, 35 (2): 182–185.
Mortazavi, A., Williams, B. A., McCue, K., Schaeffer, L., and Wold, B., 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Methods, 5 (7): 621–628.
Pawlicki, J. M., Pease, L. B., Pierce, C. M., Startz, T. P., Zhang, Y., and Smith, A. M., 2004. The effect of molluscan glue proteins on gel mechanics. Journal of Experimental Biology, 207 (7): 1127–1135.
Perezvilar, J., and Hill, R. L., 1999. The structure and assembly of secreted mucins. Journal of Biological Chemistry, 274 (45): 31751–31754.
Puchalski, S. S., Eernisse, D. J., and Johnson, C. C., 2008. The effect of sampling bias on the fossil record of chitons (Mollusca, Polyplacophora). American Malacological Bulletin, 25 (1): 87–95.
Riesgo, A., Andrade, S. C. S., Sharma, P. P., Novo, M., Pérez-Porro, A. R., Vahtera, V., González, V. L., Kawauchi, G. Y., and Giribet, G., 2012. Comparative description of ten transcriptomes of newly sequenced invertebrates and efficiency estimation of genomic sampling in non-model taxa. Frontiers in Zoology, 9: 33.
Robinson, M. D., McCarthy, D. J., and Smyth, G. K., 2010. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26 (1): 139–140.
Romkes, M., and Buch, S. C., 2014. Genotyping technologies: Application to biotransformation enzyme genetic polymorphism screening. Methods in Molecular Biology, 1105: 99–115.
Schwabe, E., 2005. A catalogue of Recent and fossil chitons (Mollusca: Polyplacophora) Addenda. Novapex, 6 (4): 89–105.
Liu, S. K., Zhang, Y., Zhou, Z. C., Waldbieser, G., Sun, F., Lu, G. G., Zhang, J. R., Jiang, Y. L., Zhang, H., Wang, X. L., Rajendran, K. V., Khoo, L., Kucuktas, H., Peatman, E., and Liu, Z. J., 2012. Efficient assembly and annotation of the transcriptome of catfish by RNA-Seq analysis of a doubled haploid homozygote. BMC Genomics, 13: 595.
Smith, A. M., 2002. The structure and function of adhesive gels from invertebrates. Integrative and Comparative Biology, 42 (6): 1164–1171.
Smith, A. M., Quick, T. J., and Peter, R. S., 1999. Differences in the composition of adhesive and non-adhesive mucus from the limpet Lottia limatula. The Biological Bulletin, 196 (1): 34–44.
Stenn, K. S., Madri, J. A., and Roll, F. J., 1979. Migrating epidermis produces AB2 collagen and requires continual collagen synthesis for movement. Nature, 277 (5693): 229–232.
Treves, K., Traub, W., Weiner, S., and Addadi, L., 2003. Aragonite formation in the chiton (Mollusca) girdle. Helvetica Chimica Acta, 86 (4): 1101–1112.
Vinther, J., and Nielsen, C., 2005. The early Cambrian Halkieria is a mollusc. Zoologica Scripta, 34 (1): 81–89.
Waite, J. H., Hansen, D. C., and Little, K. T., 1989. The glue protein of ribbed mussels (Geukensia demissa): A natural adhesive with some features of collagen. Journal of Comparative Physiology B, 159 (5): 517–525.
Wang, S., Hou, R., Bao, Z., Du, H., He, Y., Su, H., Zhang, Y., Fu, X., Jiao, W., and Li, Y., 2013. Transcriptome sequencing of Zhikong scallop (Chlamys farreri) and comparative transcriptomic analysis with Yesso scallop (Patinopecten yessoensis). PLoS One, 8 (5): e63927.
Zhou, Z. C., Dong, Y., Sun, H. J., Yang, A. F., Chen, Z., Gao, S., Jiang, J. W., Guan, X. Y., Jiang, B., and Wang, B., 2014. Transcriptome sequencing of sea cucumber (Apostichopus japonicus) and the identification of gene-associated markers. Molecular Ecology Resources, 14 (1): 127–138.
Acknowledgements
We acknowledge grant support from the National Natural Science Foundation of China (Nos. 31130054, 31472258), the AoShan Talents Program of Qingdao National Laboratory for Marine Science and Technology (No. 2015ASTP-ES02), and the Fundamental Research Funds for the Central Universities (No. 201564009).
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Dou, H., Miao, Y., Li, Y. et al. Characterization of Chiton Ischnochiton hakodadensis Foot Based on Transcriptome Sequencing. J. Ocean Univ. China 17, 632–640 (2018). https://doi.org/10.1007/s11802-018-3525-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11802-018-3525-7