Abstract
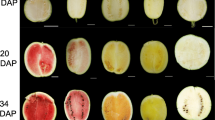
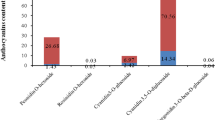
In order to screen the genes controlling watermelon rind color and luster, the experiment was carried out with yellow watermelon skin mutants as tester and green wild type watermelon as control, and transcriptome sequencing and bioinformatics analysis were done. The results show that 34.27Gb clean data were got by transcriptome sequencing. There are 261 differentially expressed genes among Y1_vs_G1, Y2_vs_G2 and Y3_vs_G3. The pathways contenting most differentially expressed genes are plant hormone signal transduction pathway, phenylpropanoid biosynthesis pathway, photosynthesis pathway, starch and sucrose metabolism pathway. 9-cis-epoxycarotenoid dioxygenase (Cla002942), alcohol dehydrogenase (Cla004992), photosystem I reaction center subunit III, chloroplastic (precursor) (Cla009181), long-chain acyl coenzyme A synthetase (Cla017341), threonine dehydratase biosynthetic (Cla018352) candidates genes were screened out.
Similar content being viewed by others
References
HUANG Shi-jie. The genetics of the watermelon rind yellow [J]. China Watermelon and Muskmelon, 1995(3): 15–16. (in Chinese)
SZAMOSI C, SOLMAZ I, SARI N, BARSONY C. Morphological characterization of Hungarian and Turkish watermelon (Citrullus lanatus (Thunb.) Matsum. et Nakai) genetic resources [J]. Genetic Resources Crop Evolution, 2009, 56(8): 1091–1105.
COCHRAN F D. Breeding cucumber for resistance to downy mildew [J]. Proceedings American Society for Horticultural Science, 1938, 35: 541–543.
LAWRENCE K P, TODD C W. Review of genes and linkage groups in cucumber [J]. Hortscience, 1990, 25(6): 605–615.
STRONG W J. Breeding experiments with the cucumber [J]. Scientia Agricola, 1931, 11: 333–346.
TKACHENKO N N. Preliminary results of a genetic investigation of the cucumber, cucumissativusL [J]. Bul Applied Plant Breed, 1935, 2(9): 311–356.
LIN Bao-ming, LIN Shi-sen. Prelimary study on the heredity of fruit length of angular sponge gourd [J]. Journal of South China Agricultural University, 2000, 21(2): 8–9. (in Chinese)
YANG Guang-hua, FAN Rong, YANG Xiao-feng, HOU Jun-liang, YUAN Shi-chen, CAO Ming, WANG Xue-lin, LI Jing-song. Construction of a highly dense genetic map using SNP and mapping of three qualitative traints in Cucumis meb [J]. Acta Horticultural Sinica, 2014, 41(5): 898–906. (in Chinese)
HOEN P A, ARIYUREK Y, THYGESEN H H, VREUGDENHI E, VOSSEN R H, MENEZES R X, BOER J M, OMMEN G J, DUNNEN J T. Deep sequencing-based expression analysis shows major advances in robustness, resolution and inter-lab portability over five microarray platforms [J]. Nucleic Acids Research, 2008, 36(21): e141.
MURAT F, XU J H, TANNIER E, ABROUK M, GUILHOT N, PONT C, MESSING J, SALSE J. Ancestral grass karyotype reconstruction unravels new mechanisms of genome shuffling as a source of plant evolution [J]. Genome Research, 2010, 20: 1545–1557.
DENG Yang-yang, LI Jian-qi, WU Song-feng, ZHU Yun-ping, CHEN Yao-wen, HE Fu-chu. Integrated NR database in protein annotation system and its localization [J]. Computer Engineering, 2006, 32(5): 71–74.
ASHBURNER M, BALL C A, BLAKE J A, BOTSTEIN D, BUTLER H, CHERRY J M, DAVIS A P, DOLINSKI K, DWIGHT S S, EPPIG J T, HARRIS M A, HILL D P, ISSEL T L, KASARSKIS A, LEWIS S, MATESE J C, RICHARDSON J E, RINGWALD M, RUBIN G M, SHERLOCK G. Gene ontology: tool for the unification of biology [J]. Nature Genetics, 2000, 25(1): 25–29.
TATUSOV R L, GALPERIN M Y, NATALE D A. The COG database: A tool for genome scale analysis of protein functions and evolution [J]. Nucleic Acids Research, 2000, 28(1): 33–36.
KANEHISA M, GOTO S, KAWASHIMA S, OKUNO Y, HATTORI A M. The KEGG resource for deciphering the genome [J]. Nucleic Acids Research, 2004, 32: D277–D280.
TRAPNELL C, WILLIAMS B A, PERTEA G, MORTAZAVI A, KWAN G, van BAREN M J, SALZBERG S L, WOLD B J, PACHTER L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation [J]. Nature Biotechnology, 2010, 28(5): 511–515.
SATOSHI I, MASATOMO K, TERUAKI T, MASAAKI N, MOTOAKI S, TOMOHIKO K, SATOSHI T, YOSHITAKA K, KAZUKO Y, KAZUO S. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in arabidopsis[J]. The Plant Journal, 2001, 27(4): 325–333.
QIN Xiao-qiang, JAN A D. Overexpression of a 9-cisepoxycarotenoid dioxygenase gene in nicotiana plumbaginifolia increases abscisic acid and phaseic acid levels and enhances drought tolerance [J]. Plant Physiology February, 2002, 128(2): 544–551.
XIA Hui, WU Shan, MA Feng-wang. Cloning and expression of two 9-cis-epoxycarotenoid dioxygenase genes during fruit development and under stress conditions from Malus [J]. Molecular Biology Reports, 2014, 41(10): 6795–6802.
LUCA D, ROBERTO B, ALEXANDER R. Photoprotective mechanisms: Carotenoids [J]. Advances in Plant Biology, 2014, 5: 393–435.
AKIHIKO Y, SAKAE K. A photoactive photosystem-II reaction-center complex lacking a chlorophyll-binding 40 kilodalton subunit from the thermophilic cyanobacterium Synechococcus sp [J]. Biochimica et Biophysica Acta, 1984, 765(2): 118–124.
ROSALIND A C, TAL M L, CYNTHIA G. Do long-chain acyl-coa synthetasesregulate fatty acid entry into synthetic versus degradative pathways [J]. Nutrition, 2002, 132(8): 2123–2126.
SOUPENE E, KUYPERS F A. Mammalian long-chain acyl-coa synthetases [J]. Experimental Biology Medicine, 2008, 233(5): 507–521.
MOCKEL B, EGGELING L, SAHM H. Functional and structural analyses of threonine dehydratase from Corynebacterium glutamicum [J]. Bacteriol, 1992, 174: 8065–8072.
SHARMA R, KESHARI D, SINGH K S, YADAV S, SINGH S K. MRA_1571 is required for isoleucine biosynthesis and improves mycobacterium tuberculosis H37Ra survival under stress [J]. Scientific Reports, 2016, 6: 27997.
DONG Xun-yan, ZHAO Yue, HU Jin-yu, WANG Xiao-yuan, LI Ye. Attenuating l -lysine production by deletion of ddh and lysE and their effect on L-threonine and L-isoleucine production in Corynebacterium glutamicum [J]. Enzyme and Microbial Technology, 2016, 93/94: 70–78.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Foundation item: Project(31260476) supported by the National Natural Science Foundation of China
Rights and permissions
About this article
Cite this article
Yang, Kk., Liang, Zh. & Wu, Cj. Analysis on differentially expressed genes in watermelon rind color based on RNA–Seq. J. Cent. South Univ. 23, 2818–2826 (2016). https://doi.org/10.1007/s11771-016-3345-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11771-016-3345-0