Abstract
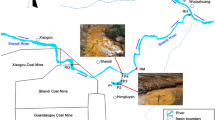
To reveal the impact of mining on bacterial ecology around mining area, bacterial community and geochemical characteristics about Dabaoshan Mine (Guangdong Province, China) were studied. By amplified ribosomal DNA restriction analysis and phylogenetic analysis, it is found that mining pollution greatly impacts the bacterial ecology and makes the habitat type of polluted environments close to acid mine drainage (AMD) ecology. The polluted environment is acidified so greatly that neutrophil and alkaliphilic microbes are massively dead and decomposed. It provided organic matters that can make Acidiphilium sp. rapidly grow and become the most bacterial species in this niche. Furthermore, Acidithiobacillus ferrooxidans and Leptospirillum sp. are also present in this niche. The amount of Leptospirillum sp. is far more than that of Acidithiobacillus ferrooxidans, which indicates that the concentration of toxic ions is very high. The conclusions of biogeochemical analysis and microbiological monitor are identical. Moreover, because the growth of Acidithiobacillus ferrooxidans and Leptospirillum sp. depends on ferrous iron or inorganic redox sulfur compounds which can be supplied by continual AMD, their presence indicates that AMD still flows into the site. And the area is closer to the outfalls of AMD, their biomasses would be more. So the distinction of their biomasses among different areas can help us to find the effluent route of AMD.
Similar content being viewed by others
References
JOHNSON D B, HALLBERG K B. The microbiology of acidic mine waters [J]. Research in Microbiology, 2003, 154(7): 466–473.
BAKER B J, BANFIELD J F. Microbial communities in acid mine drainage [J]. Fems Microbiology Ecology, 2003, 44(2): 139–152.
OKABAYASHI A, WAKAI S, KANAO T, SUGIO T, KAMIMURA K. Diversity of 16S ribosomal DNA-defined bacterial population in acid rock drainage from Japanese pyrite mine [J]. Journal of Bioscience and Bioengineering, 2005, 100(6): 644–652.
YANG Yu, SHI Wu-yang, WAN Min-xi, ZHANG Yan-fei, ZOU Li-hong, HUANG Ju-fang, QIU Guan-zhou, LIU Xue-duan. Diversity of bacterial communities in acid mine drainage from the Shen-bu copper mine, Gansu Province, China [J]. Electronic Journal of Biotechnology, 2008, 11(1): 108–117.
ZHOU Hong-bo, LIU Xi, FU Bo, QIU Guan-zhou, HUO Qiang, ZENG Wei-min, LIU Jian-she, CHEN Xin-hua. Isolation and characterization of acidithiobacillus caldus from several typical environments in China [J]. Journal of Central South University of Technology, 2007, 14(2): 163–169.
YIN Hua-qun, QIU Guan-zhou, WANG Dian-zuo, CAO Lin-hui, DAI Zhi-min, WANG Jie-wei, LIU Xue-duan. Comparison of microbial communities in three different mine drainages and their bioleaching efficiencies to low grade of chalcopyrite [J]. Journal of Central South University of Technology, 2007, 14(4): 460–466.
HURT R A, QIU X Y, WU L Y, ROH Y, PALUMBO A V, TIEDJE J M, ZHOU J Z. Simultaneous recovery of RNA and DNA from soils and sediments [J]. Applied and Environmental Microbiology, 2001, 67(10): 4495–4503.
MARCHESI J R, SATO T, WEIGHTMAN A J, MARTIN T A, FRY J C, HIOM S J, WADE W G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA [J]. Applied and Environmental Microbiology, 1998, 64(2): 795–799.
SMITH S W, OVERBEEK R., WOESE C R., GILBERT W, GILLEVET P M. The genetic data environment an expandable gui for multiple sequence-analysis [J]. Computer Applications in the Biosciences, 1994, 10(6): 671–675.
ZHOU J Z, XIA B C, TREVES D S, WU L Y, MARSH T L, O’NEILL R V, PALUMBO A V, TIEDJE J M. Spatial and resource factors influencing high microbial diversity in soil [J]. Applied and Environmental Microbiology, 2002, 68(1): 326–334.
RAVENSCHLAG K, SAHM K, PERNTHALER J, AMANN R. High bacterial diversity in permanently cold marine sediments [J]. Applied and Environmental Microbiology, 1999, 65(9): 3982–3989.
STACH J E M, MALDONADO L A, MASSON D G, WARD A C, GOODFELLOW M, BULL A T. Statistical approaches for estimating actinobacterial diverity in marine sediments [J]. Applied and Environmental Microbiology, 2003, 69(10): 6189–6200.
DRUSCHEL G K, BAKER B J, GIHRING T M, BANFIELD J F. Acid mine drainage biogeochemistry at Iron Mountain, California [J]. Geochemical Transactions, 2004, 5(2): 13–32.
PECCIA J, MARCHAND E A, SILVERSTEIN J, HERNANDEZ M. Development and application of small-subunit rRNA probes for assessment of selected Thiobacillus species and members of the genus Acidiphilium [J]. Applied and Environmental Microbiology, 2000, 66(7): 3065–3072.
GONZALEZ-TORIL E, LLOBET-BROSSA E, CASAMAYOR E O, AMANN R, AMILS R. Microbial ecology of an extreme acidic environment, the Tinto River [J]. Applied and Environmental Microbiology, 2003, 69(8): 4853–4865.
JOHNSON D B. Selective solid media for isolating and enumerating acidophilic bacteria [J]. Journal of Microbiological Methods, 1995, 23(2): 205–218.
KUSEL K, DORSCH T, ACKER G, STACKEBRANDT E. Microbial reduction of Fe(III) in acidic sediments: Isolation of Acidiphilium cryptum JF-5 capable of coupling the reduction of Fe(III) to the oxidation of glucose [J]. Applied and Environmental Microbiology, 1999, 65(8): 3633–3640.
JOHNSON D B, ROLFE S, HALLBERG K B, IVERSEN E. Isolation and phylogenetic characterization of acidophilic microorganisms indigenous to acidic drainage waters at an abandoned Norwegian copper mine [J]. Environmental Microbiology, 2001, 3(10): 630–637.
ROHWERDER T, GEHRKE T, KINZLER K, SAND W. Bioleaching review part A: Progress in bioleaching: fundamentals and mechanisms of bacterial metal sulfide oxidation [J]. Applied Microbiology and Biotechnology, 2003, 63(3): 239–248.
FOWLER T A, HOLMES P R, CRUNDWELL F K. Mechanism of pyrite dissolution in the presence of Thiobacillus ferrooxidans [J]. Applied and Environmental Microbiology, 1999, 65(7): 2987–2993.
BOND P L, SMRIGA S P, BANFIELD J F. Phylogeny of microorganisms populating a thick, subaerial, predominantly lithotrophic biofilm at an extreme acid mine drainage site [J]. Applied and Environmental Microbiology, 2000, 66(9): 3842–3849.
HIPPE H. Leptospirillum gen. nov (ex Markosyan 1972), nom. rev., including Leptospirillum ferrooxidans sp nov (ex Markosyan 1972), nom. rev. and Leptospirillum thermoferrooxidans sp nov (Golovacheva et al. 1992) [J]. International Journal of Systematic and Evolutionary Microbiology, 2000, 50: 501–503.
CORAM N J, RAWLINGS D E. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptosphillum ferriphilum sp nov dominates South African commercial biooxidation tanks that operate at 40°C [J]. Applied and Environmental Microbiology, 2002, 68(2): 838–845.
YANG Yu, WAN Min-xi, SHI Wu-yang, PENG Hong, QIU Guang-zhou, ZHOU Ji-zhong, LIU Xue-duan. Bacterial diversity and community structure in acid mine drainage from Dabaoshan Mine, China [J]. Aquatic Microbial Ecology, 2007, 47(2): 141–151.
Author information
Authors and Affiliations
Corresponding author
Additional information
Foundation item: Project(50621063) supported by the Science Fund for Creative Research Groups of China; Project(2004CB619201) supported by the Major State Basic Research Development Program of China
Rights and permissions
About this article
Cite this article
Wan, Mx., Yang, Y., Qiu, Gz. et al. Acidophilic bacterial community reflecting pollution level of sulphide mine impacted by acid mine drainage. J. Cent. South Univ. Technol. 16, 223–229 (2009). https://doi.org/10.1007/s11771-009-0038-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11771-009-0038-y