Abstract
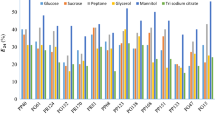
A total of seventy-one bacteria were isolated from crude oil-contaminated soil samples. The isolates were screened for biosurfactant production by applying various methods, including determination of hemolytic, lipolytic, and esterolytic activities, oil displacement, parafilm M, drop-collapse, CTAB-methylene blue agar tests, determination of emulsification index (E24) and reduction in surface tension. Thirteen isolates were selected and further subjected to molecular identification. 16S rRNA gene sequencing analysis indicated that the isolates belong to Pseudomonas (Pseudomonas sp., P. balearica, and P. aeruginosa), Acinetobacter (A. radioresistens, A. baumanni), Bacillus (Bacillus sp.) and Priestia (Priestia sp.) genera. The largest spreading zone diameter (6.5 cm) and the highest enzymatic activity index (EAI) for lipolytic (5.4) and esterolytic (2.7) activities were demonstrated by P. aeruginosa strain TBA-B58. Surface tension analysis by the Du Noüy tensiometer confirmed that the glycolipid-type biosurfactant obtained from P. aeruginosa strain TBA-B58 achieved the highest reduction in surface tension with a value of 31.5 ± 0.5 mN m−1. Notable E24 values, superior to the synthetic surfactant sodium dodecyl sulfate, were recorded by the cell-free supernatants obtained from A. baumanni TBA-B45 (66%) and TBA-B56 (65%) against toluene and benzene, respectively. Based on the results of the present study, two biosurfactant/bioemulsifier-producing strains belonging to Pseudomonas and Acinetobacter genera demonstrated significant surface and interfacial activities, which might find applications in the bioremediation of crude oil hydrocarbons.


Similar content being viewed by others
Data availability
The sequencing data that support the molecular findings of this study are openly available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
Abbreviations
- EAI:
-
Enzymatic Activity Index
- E24 :
-
Emulsification Index
- GPS:
-
Global Positioning System
- NA:
-
Nutrient Agar
- LB:
-
Luria Bertani
- BPM:
-
Biosurfactant Production Medium
- MSM:
-
Mineral Salt Medium
- CFS:
-
Cell-free Supernatant
- SDS:
-
Sodium Dodecyl Sulfate
- CTAB-MB:
-
Cetyltrimethylammonium Bromide-Methylene Blue
- PCR:
-
Polymerase Chain Reaction
- NCBI:
-
National Center for Biotechnology Information
- bp:
-
Base pair
- ODD:
-
Oil Displacement Diameter
- ODA:
-
Oil Displacement Area
- PAHs:
-
Polycyclic Aromatic Hydrocarbons
- HA:
-
Hemolytic Activity
- LA:
-
Lipolytic Activity
- EA:
-
Esterolytic Activity
- BA:
-
CTAB-MB Agar
- DC:
-
Drop Collapse Assay
- PM:
-
Parafilm M Test
- OD:
-
Oil Displacement Assay
- EF:
-
Emulsion Formation Capacity
References
Al-Saleh E, Drobiova H, Obuekwe C (2009) Predominant culturable crude oil-degrading bacteria in the coast of Kuwait. Int Biodeter Biodegr 63(4):400–406. https://doi.org/10.1016/j.ibiod.2008.11.004
Anburajan L, Meena B, Raghavan RV, Shridhar D, Joseph TC, Vinithkumar NV, Dharani G, Dheenan PS, Kirubagaran R (2015) Heterologous expression, purification, and phylogenetic analysis of oil-degrading biosurfactant biosynthesis genes from the marine sponge-associated Bacillus licheniformis NIOT-06. Bioprocess Biosyst Eng 38:1009–1018. https://doi.org/10.1007/s00449-015-1359-x
Asemoloye MD, Ahmad R, Jonathan SG (2017) Synergistic action of rhizospheric fungi with Megathyrsus maximus root speeds up hydrocarbon degradation kinetics in oil-polluted soil. Chemosphere 187:1–10. https://doi.org/10.1016/j.chemosphere.2017.07.158
Chen Q, Li J, Liu M, Sun H, Bao M (2017) Study on the biodegradation of crude oil by free and immobilized bacterial consortium in marine environment. PLoS One 12(3):e0174445. https://doi.org/10.1371/journal.pone.0174445
Colla LM, Rizzardi J, Pinto MH, Reinehr CO, Bertolin TE, Costa JAV (2010) Simultaneous production of lipases and biosurfactants by submerged and solid-state bioprocesses. Biores Technol 101:8308–8314. https://doi.org/10.1016/j.biortech.2010.05.086
Dave D, Ghaly AE (2011) Remediation technologies for marine oil spills: a critical review and comparative analysis. Am J Environ Sci 7(5):423–440
Elumalai P, Parthipan P, Huang M, Muthukumar B, Cheng L, Govarthanan M, Rajasekar A (2021) Enhanced biodegradation of hydrophobic organic pollutants by the bacterial consortium: Impact of enzymes and biosurfactants. Environ Pollut 289:117956. https://doi.org/10.1016/j.envpol.2021.117956
Fatahinia M, Poormohamadi F, Mahmoudabadi AZ (2015) Comparative study of esterase and hemolytic activities in clinically important Candida species, isolated from oral cavity of diabetic and non-diabetic individuals. Jundishapur J Microbiol 8(3):e20893. https://doi.org/10.5812/jjm.20893
Felix AKN, Martins JJL, Almeida JGL, Giro MEA, Cavalcante KF, Melo VMM, Pessoa ODL, Rocha MVP, Gonçalves LRB, de Santiago Aguiar RS (2019) Purification and characterization of a biosurfactant produced by Bacillus subtilis in cashew apple juice and its application in the remediation of oil-contaminated soil. Colloids Surf B Biointerfaces 175:256–263. https://doi.org/10.1016/j.colsurfb.2018.11.062
Felsenstein J (1985) Confidence limits on phylogenies: An approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Galieriková A, Materna M (2020) World seaborne trade with oil: one of main cause for oil spills? Transp Res Proc 44:297–304. https://doi.org/10.1016/j.trpro.2020.02.039
Gudiña EJ, Pereira JFB, Rodrigues LR, Coutinho JAP, Teixeira JA (2012) Isolation and study of microorganisms from oil samples for application in microbial enhanced oil recovery. Int Biodeterior Biodegradation 68:56–64. https://doi.org/10.1016/j.ibiod.2012.01.001
Gupta B, Puri S, Thakur IS, Kaur J (2020) Enhanced pyrene degradation by a biosurfactant producing Acinetobacter baumannii BJ5: growth kinetics, toxicity and substrate inhibition studies. Environ Technol Innov 19:100804. https://doi.org/10.1016/j.eti.2020.100804
Imam A, Suman SK, Ghosh D, Kanaujia PK (2019) Analytical approaches used in monitoring the bioremediation of hydrocarbons in petroleum-contaminated soil and sludge. TrAC Trends Anal Chem 118:50–64. https://doi.org/10.1016/j.trac.2019.05.023
Janek T, Mirończuk AM, Rymowicz W, Dobrowolski A (2020) High-yield expression of extracellular lipase from Yarrowia lipolytica and its interactions with lipopeptide biosurfactants: a biophysical approach. Arch Biochem Biophys 689:108475. https://doi.org/10.1016/j.abb.2020.108475
Jimoh AA, Senbadejo TY, Adeleke R, Lin J (2021) Development and genetic engineering of hyper-producing microbial strains for improved synthesis of biosurfactants. Mol Biotechnol 63:267–288. https://doi.org/10.1007/s12033-021-00302-1
Jimoh AA, Ikhimiukor OO, Adeleke R (2022) Prospects in the bioremediation of petroleum hydrocarbon contaminants from hypersaline environments: a review. Environ Sci Pollut Res 29:35615–35642. https://doi.org/10.1007/s11356-022-19299-4
Kreling NE, Simon V, Fagundes VD, Thomé A, Colla LM (2020) Simultaneous production of lipases and biosurfactants in solid-state fermentation and use in bioremediation. J Environ Eng 146(9):04020105. https://doi.org/10.1061/(ASCE)EE.1943-7870.0001785
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kumari S, Regar RK, Manickam N (2018) Improved polycyclic aromatic hydrocarbon degradation in a crude oil by individual and a consortium of bacteria. Bioresour Technol 254:174–179. https://doi.org/10.1016/j.biortech.2018.01.075
Mohanram R, Jagtap C, Kumar P (2016) Isolation, screening, and characterization of surface-active agent-producing, oil-degrading marine bacteria of Mumbai Harbor. Mar Pollut Bull 105(1):131–138. https://doi.org/10.1016/j.marpolbul.2016.02.040
Morikawa M, Hirata Y, Imanaka T (2000) A study on the structure-function relationship of lipopeptide biosurfactants. BBA-Mol Cell Biol L 1488(3):211–218. https://doi.org/10.1016/S1388-1981(00)00124-4
Mujumdar S, Joshi P, Karve N (2019) Production, characterization, and applications of bioemulsifiers (BE) and biosurfactants (BS) produced by Acinetobacter spp.: a review. J Basic Microbiol 59(3):277–87. https://doi.org/10.1002/jobm.201800364
Muneeswari R, Swathi KV, Sekaran G, Ramani K (2021) Microbial-induced biosurfactant-mediated biocatalytic approach for the bioremediation of simulated marine oil spill. Int J Environ Sci Technol 19:341–354. https://doi.org/10.1007/s13762-020-03086-0
Muthukamalam S, Sivagangavathi S, Dhrishya D, Sudha Rani S (2017) Characterization of dioxygenases and biosurfactants produced by crude oil degrading soil bacteria. Braz J Microbiol 48:637–647
Ndimele PE, Saba AO, Ojo DO, Ndimele CC, Anetekhai MA, Erondu ES (2018) Remediation of crude oil spillage. In: Ndimele PE (ed) The political ecology of oil and gas activities in the Nigerian aquatic ecosystem. Academic Press, Cambridge, pp 369–384. https://doi.org/10.1016/B978-0-12-809399-3.00024-0
Nejad YS, Jaafarzadeh N, Ahmadi M, Abtahi M, Ghafari S, Jorfi S (2020) Remediation of oily sludge wastes using biosurfactant produced by bacterial isolate Pseudomonas balearica strain Z8. J Environ Health Sci Eng 18:531–539. https://doi.org/10.1007/s40201-020-00480-1
Okoh E, Yelebe ZR, Oruabena B, Nelson ES, Indiamaowei OP (2020) Clean-up of crude oil-contaminated soils: bioremediation option. Int J Environ Sci Technol 17:1185–1198. https://doi.org/10.1007/s13762-019-02605-y
Oliveira JS, Araújo WJ, Figueiredo RM, Silva-Portela RCB, Guerra ADB, Araújo SCS, Minnicelli C, Carlos AC, Tereza A, Vasconcelos RD, Freitas AT, Agnez-Lima LF (2017) Biogeographical distribution analysis of hydrocarbon degrading and biosurfactant producing genes suggests that near-equatorial biomes have higher abundance of genes with potential for bioremediation. BMC Microbiol 17:168. https://doi.org/10.1186/s12866-017-1077-4
Pajiar N, Sachan SG, Sachan A (2017) Biosurfactants: a multifunctional microbial metabolite. In: Kalia V (ed) Microbial applications, vol 2. Springer, Cham, pp 213–229. https://doi.org/10.1007/978-3-319-52669-0_12
Parthipan P, Cheng L, Dhandapani P, Elumalai P, Huang M, Rajasekar A (2022) Impact of biosurfactant and iron nanoparticles on biodegradation of polyaromatic hydrocarbons (PAHs). Environ Pollut 3:119384. https://doi.org/10.1016/j.envpol.2022.119384
Phan CW, Abu Bakar NF, Hamzah A (2013) A comparative study on biosurfactant activity of crude oil-degrading bacteria and its correlation to total petroleum hydrocarbon degradation. Bioremediat J 17(4):240–251. https://doi.org/10.1080/10889868.2013.827621
Prince RC, Amande TJ, McGenity TJ (2019) Prokaryotic hydrocarbon degraders. In: McGenity T (ed) Taxonomy, genomics and ecophysiology of hydrocarbon-degrading microbes. Handbook of hydrocarbon and lipid microbiology. Springer, Cham, pp 1–39. https://doi.org/10.1007/978-3-030-14796-9_15
Rahman PKSM, Pasirayi G, Auger V, Ali Z (2010) Production of rhamnolipid biosurfactants by Pseudomonas aeruginosa DS10-129 in a microfluidic bioreactor. Biotechnol Appl Biochem 55(1):45–52. https://doi.org/10.1042/BA20090277
Roy A, Dutta A, Pal S, Gupta A, Sarkar J, Chatterjee A, Saha A, Sarkar P, Sar P, Kazy SK (2018) Biostimulation and bioaugmentation of native microbial community accelerated bioremediation of oil refinery sludge. Bioresour Technol 253:22–32. https://doi.org/10.1016/j.biortech.2018.01.004
Sah D, Rai JP, Ghosh A, Chakraborty M (2022) A review on biosurfactant producing bacteria for remediation of petroleum contaminated soils. 3 Biotech 12(9):1–31. https://doi.org/10.1007/s13205-022-03277-1
Saitou N, Nei M (1987) The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Sarkar J, Roy A, Sar P, Kazy SK (2020) Accelerated bioremediation of petroleum refinery sludge through biostimulation and bioaugmentation of native microbiome. In: Shah MP, Rodriguez-Couto S, Sengor SS (eds) Emerging technologies in environmental bioremediation. Elsevier, Amsterdam, pp 23–65. https://doi.org/10.1016/B978-0-12-819860-5.00002-X
Satpute SK, Bhawsar BD, Dhakephalkar PK, Chopade BA (2008) Assessment of different screening methods for selecting biosurfactant producing marine bacteria. Indian J Mar Sci 37(3):243–250 (http://nopr.niscair.res.in/handle/123456789/2045)
Sekhon KK, Khanna S, Cameotra SS (2011) Enhanced biosurfactant production through cloning of three genes and role of esterase in biosurfactant release. Microb Cell Fact 10:49. https://doi.org/10.1186/1475-2859-10-49
Sekhon KK, Khanna S, Cameotra SS (2012) Biosurfactant production and potential correlation with esterase activity. J Pet Environ Biotechnol 3:7–16. https://doi.org/10.4172/2157-7463.1000133
Sharma A, Sharma T, Sharma T, Sharma S, Kanwar SS (2019) Role of microbial hydrolases in bioremediation. In: Kumar A, Sharma S (eds) Microbes and enzymes in soil health and bioremediation. Microorganisms for sustainability, vol 16. Springer, Singapore, pp 149–164. https://doi.org/10.1007/978-981-13-9117-0_7
Shatila F, Uyar E, Yalçın HT (2021) Screening of biosurfactant production by Yarrowia lipolytica strains and evaluation of their antibiofilm and anti-adhesive activities against Salmonella enterica ser. Enteritidis Biofilms Microbiol 90:839–847. https://doi.org/10.1134/S002626172201012X
Siles JA, García-Sánchez M (2018) Microbial dynamics during the bioremediation of petroleum hydrocarbon-contaminated soils through biostimulation: an overview. In: Prasad R, Aranda E (eds) Approaches in bioremediation- the new era of environmental microbiology and nanobiotechnology. Springer, Cham, pp 115–34. https://doi.org/10.1007/978-3-030-02369-0_7
Singh SK, Sachan A (2022) A review on biotransformation of polyaromatic hydrocarbons mediated by biosurfactant producing bacteria. Petrol Sci Technol 14:1–21. https://doi.org/10.1080/10916466.2022.2042558
Singh P, Ravindran S, Patil Y (2021) Biosurfactant enhanced sustainable remediation of petroleum contaminated soil. In: Sarma H, Prasad MNV (eds) Biosurfactants for a sustainable future: production and applications in the environment and biomedicine. Wiley, Hoboken, pp 119–138. https://doi.org/10.1002/9781119671022.ch6
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526. https://doi.org/10.1093/oxfordjournals.molbev.a040023
Trindade M, Sithole N, Kubicki S, Thies S, Burger A (2021) Screening strategies for biosurfactant discovery. In: Scheper T (ed) Advances in biochemical engineering/biotechnology. Springer, Berlin, pp 1–36. https://doi.org/10.1007/10_2021_174
Uyar E, Sağlam Ö (2021) Isolation, screening and molecular characterization of biosurfactant producing bacteria from soil samples of auto repair shops. Arch Microbiol 203:4929–4939. https://doi.org/10.1007/s00203-021-02483-4
Walter V, Syldatk C, Hausman R (2010) Screening concepts for the isolation of biosurfactant producing microorganisms. In: Sen R (ed) Biosurfactants. Advances in experimental medicine and biology, vol 672. Springer, New York, pp 1–13. https://doi.org/10.1007/978-1-4419-5979-9_1
Wang H, Jiang R, Kong D, Liu Z, Wu X, Xu J, Li Y (2020) Transmembrane transport of polycyclic aromatic hydrocarbons by bacteria and functional regulation of membrane proteins. Front Environ Sci Eng 14(1):9. https://doi.org/10.1007/s11783-019-1188-2
Wu Q, Zhi Y, Xu Y (2019) Systematically engineering the biosynthesis of a green biosurfactant surfactin by Bacillus subtilis 168. Metab Eng 52:87–97. https://doi.org/10.1016/j.ymben.2018.11.004
Yadav AK, Manna S, Pandiyan K, Singh A, Kumar M, Chakdar H, Kashyap PL, Srivastava AK (2016) Isolation and characterization of biosurfactant producing Bacillus sp. from diesel fuel-contaminated site. Microbiology 85(1):56–62. https://doi.org/10.1134/S0026261716010161
Yalçın HT, Fındık B, Terzi Y, Uyar E, Shatila F (2021) Isolation and molecular identification of industrially important enzyme producer yeasts from tree barks and fruits. Arch Microbiol 203(3):1079–1088. https://doi.org/10.1007/s00203-020-02104-6
Youssef N, Duncan K, Nagle D (2004) Comparison of methods to detect biosurfactant production by diverse microorganisms. J Microbiol Methods 56:339–347. https://doi.org/10.1016/j.mimet.2003.11.001
Zhang R, Liu Y, Huang X, Xu M, Liu R, Zong W (2018) Interaction of a digestive protease, Candida rugosa lipase, with three surfactants investigated by spectroscopy, molecular docking and enzyme activity assay. Sci Total Environ 622:306–315. https://doi.org/10.1016/j.scitotenv.2017.11.305
Zhang X, Kong D, Liu X, Xie H, Lou X, Zeng C (2021) Combined microbial degradation of crude oil under alkaline conditions by Acinetobacter baumannii and Talaromyces sp. Chemosphere 273:129666. https://doi.org/10.1016/j.chemosphere.2021.129666
Zou C, Wang M, Xing Y, Lan G, Ge T, Yan X, Gu T (2014) Characterization and optimization of biosurfactants produced by Acinetobacter baylyi ZJ2 isolated from crude oil-contaminated soil sample toward microbial enhanced oil recovery applications. Biochem Eng J 90:49–58. https://doi.org/10.1016/j.bej.2014.05.007
Acknowledgements
This study was partially supported by the Scientific Research Council of Harran University (HUBAP, grant no:19264).
Funding
This study was partially supported by the Scientific Research Council of Harran University (HUBAP, grant no:19264).
Author information
Authors and Affiliations
Contributions
EU conceived and designed the study, performed the research, analyzed the data, wrote the paper. TA performed the research except for the surface tension measurements. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent for publication
The authors “Ebru Uyar and Tuba Avcı” have approved the manuscript and agreed with the submission of the manuscript. They also declare that the paper titled “Screening and molecular identification of biosurfactant/bioemulsifier producing bacteria from crude oil contaminated soils samples” has not been published or being considered for publication elsewhere.
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uyar, E., Avcı, T. Screening and molecular identification of biosurfactant/bioemulsifier producing bacteria from crude oil contaminated soils samples. Biologia 78, 2179–2193 (2023). https://doi.org/10.1007/s11756-023-01330-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-023-01330-9