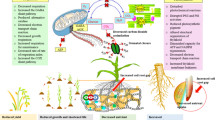
Abstract
Among all the cereals, wheat is an essential grain and food crop that provides a significant portion of the daily nutritional requirements for humans. To find the drought tolerant and drought sensitive genotypes, we employed fifty-three wheat genotypes, of which twenty-four genotypes were chosen based on physiological and biochemical criteria such as RWC, proline, and chlorophyll content. With the help of semi-quantitative RT-PCR and quantitative RT-PCR protocol, expression of dehydrin gene (WCOR410) and C-repeat binding factor genes such as CBF3, CBF4, CBF9, CBF5, CBF15, and 3H9 genes were studied in drought tolerant (C 306 and HD 2888) and sensitive genotypes (RAJ1555 and GW273) at the pre-anthesis stage. C306 and HD2888 genotypes identifies as a drought tolerant whereas RAJ1555 and GW273 genotypes shows less tolerance for drought stress condition. The majority of CBF genes were found to be up-regulated in some genotypes and down-regulated in others when exposed to water stress. In our study, we observed that genotype C306 had the highest amount of up-regulation when exposed to dehydration stress. These findings also show that the molecular response of plants to dehydration changes over time as a result of a combination of internal and external stressors. In the future, the identified tolerant variety could be used in wheat breeding for drought tolerant traits.


Similar content being viewed by others
Abbreviations
- ABA:
-
Abscisic acid
- ABRE:
-
ABA-responsive elements
- ANOVA:
-
Analysis of variance
- AP2:
-
Apetala2
- bHLH:
-
Basic helix–loop–helix
- bZIP:
-
Basic leucine zipper
- CBF:
-
C-repeat binding factor
- Ct:
-
Comparative threshold cycle
- DDF:
-
Dwarf and delayed flowering
- DREB:
-
Dehydration-responsive element binding
- EREBPs:
-
Ethylene-responsive element binding proteins
- MYB:
-
Myeloblastosis
- NLS:
-
Nuclear localization signal
- PCR:
-
Polymerase chain reaction
- RBD:
-
Random block design
- ROS:
-
Reactive oxidation species
- RWC:
-
Relative water content
- SPAD:
-
Soil plant analytical development
- SWC:
-
Soil water content
- TFs:
-
Transcription factors
References
Bajaj S, Targolli J, Liu LF, Ho DTH, Wu R (1999) Transgenic approaches to increase dehydration-stress tolerance in plants. Mol Breed 5:493–503. https://doi.org/10.1023/A:1009660413133
Bates LS, Waldren RP, Tear ID (1973) Rapid determination of free proline for water-stress studies. Plant Soil 39:205–207. https://doi.org/10.1007/BF00018060
Blum A (1988) Plant Breeding for Stress Environments. CRC Press Inc., Boca Raton, Florida, USA. https://doi.org/10.1201/9781351075718
Chen TH, Murata N (2002) Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Curr Opin Plant Biol 5:250–257. https://doi.org/10.1016/s1369-5266(02)00255-8
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetics perspectives on cross-talk and specificity in abiotic stress signaling in plants. J Exp Bot 55:225–236
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Annu Rev Biochem 162:156–159. https://doi.org/10.1006/abio.1987.9999
Elisabet B (1997) Plant responses to water deficit. Trends Plant Sci 2:48–54. https://doi.org/10.1007/BF00024008
Farquhar GD, Von CS, Berry JA (1980) A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 149:78–90. https://doi.org/10.1007/BF00386231
Hsieh TH, Lee JT, Charng YY, Chan MT (2002) Tomato plants ectopically expressing Arabidopsis CBF1 show enhanced resistance to water deficit stress. Plant Physiol 130:618–626. https://doi.org/10.1104/pp.006783
Kasuga M, Liu Q, Miura S, Yamaguchi-Shinozaki K, Shinozaki K (1999) Improving plant drought, salt and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat Biotechnol 17:287–291. https://doi.org/10.1038/7036
Kidokoro S, Watanabe K, Ohori T, Moriwaki T, Maruyama K et al (2015) Soybean DREB1/CBF-type transcription factors function in heat and drought as well as cold stress-responsive gene expression. Plant J 81:505–518. https://doi.org/10.1111/tpj.12746
Knox AK, Li C, Vagujfalvi A, Galiba G, Stockinger EJ, Dubcovsky J (2008) Identification of candidate CBF genes for the frost tolerance locus Fr-Am2 in Triticum monococcum. Plant Mol Biol 67:257–270. https://doi.org/10.1007/s11103-008-9316-6
Korir P, Nyabundi J, Kimurto P (2006) Genotypic response of common bean (Phaseolus vulgaris. L). to moisture stress condition in Kenya. Asian J Plant Sci 5:24–32. https://doi.org/10.3923/ajps.2006.24.32
Leonardis AMD, Marone D, Mazzucotelli E, Neffar F, Rizza F et al (2007) Durum wheat genes up-regulated in the early phases of cold stress are modulated by drought in a developmental and genotype dependent manner. Plant Sci 172:1005–1016. https://doi.org/10.1016/j.plantsci.2007.02.002
Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S et al (1998) Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought and low temperature responsive gene expression, respectively, in Arabidopsis. Plant Cell 10:1391–1406. https://doi.org/10.1105/tpc.10.8.1391
Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2– ∆∆Ct Method. Methods 25:402–408
Mare C, Mazzucotelli E, Crosatti C, Francia E, Stanca AM et al (2004) Hv-WRKY38: a new transcription factor involved in cold and drought-response in barley. Plant Mol Biol 55:399–416. https://doi.org/10.1007/s11103-004-0906-7
Molinari HBC, Marur CJ, Daros E, Freitas DMK, Portela DJFR et al (2007) Evaluation of the stress inducible production of proline in transgenic sugarcane (Saccharum spp.): osmotic adjustment, chlorophyll fluorescence and oxidative stress. Physiol Plant 130:218–229. https://doi.org/10.1111/j.1399-3054.2007.00909.x
Niu X, Luo T, Zhao H, SuY W, Ji, Li H (2020) Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene Gene. doi: https://doi.org/10.1016/j.gene.2020.144514
Oh SJ, Song SI, Kim YS, Jang HJ, Kim SY et al (2005) Arabidopsis CBF3/DREB1A and ABF3 in transgenic rice increased tolerance to abiotic stress without stunting growth. Plant Physiol 138:341–351. https://doi.org/10.1104/pp.104.059147
Pandey P, Irulappan V, Bagavathiannan MV, Muthappa SK (2017) Impact of Combined Abiotic and Biotic Stresses on Plant Growth and Avenues for Crop Improvement by Exploiting Physio-morphological Traits. Front Plant Sci 8:537. https://doi.org/10.3389/fpls.2017.00537
Chandrasekar V, Sairam RK, Srivastava GC (2000) Physiological and Biochemical Responses of Hexaploid and Tetraploid Wheat to Drought Stress. J Agron Crop Sci 074:108–116. https://doi.org/10.1046/j.1439-037x.2000.00430.x
Rampino P, Pataleo S, Gerardi C, Mita G, Perrotta C (2006) Drought stress response in wheat: physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ 29:2143–2152. https://doi.org/10.1111/j.1365-3040.2006.01588.x
Sairam RK (1994) Effect of Moisture Stress on Physiological Activities of Two Contrasting Wheat Genotypes. Indian J Exp Biol 32:594–597
Slatyer RO, Barrs HD (1965) Modifications to the relative turgidity technique with notes on its significance as an index of the internal water status of leaves. Methodology of plant Ecophysiol (E.E. Eskardt, ed.) UNESCO, Paris
Sakuma Y, Maruyama K, Osakabe Y, Qin F, Seki M et al (2006) Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18:1292–1309. https://doi.org/10.1105/tpc.105.035881
Sankar C, Abdul JP, Manivannan A, Kishore KR, Somasundaram R, Selvam P (2007) Effect of paclobutrazol on water stress amelioration through antioxidants and free radical scavenging enzymes in Arachis hypogaea L. Colloids Surf B Biointerfaces 60:229–235. https://doi.org/10.1016/j.colsurfb.2007.06.016
Shen YG, Zhang WK, He SJ, Zhang JS, Liu Q, Chen SY (2003) An EREBP/AP2 type protein in Triticum aestivum was a DRE binding transcription factor induced by cold, dehydration, and ABA stress. Theor Appl Genet 106:923–930. https://doi.org/10.1007/s00122-002-1131-x
Sinclair TR, Ludlow MM (1985) Who taught plants thermodynamics? The unfulfilled potential of plant water potential. Aust J Plant Physiol 12:213–217. https://doi.org/10.1071/PP9850213
Stocker O (1929) Das Wasserdefizit von Gefässpflanzen in verschiedenen Klimazonen. Planta 7:382–387. https://doi.org/10.1007/BF01916035
Sutton F, Chen XG, Don K (2009) genes of the Fr-A2 allele are differentially regulated between long-term cold acclimated crown tissue of freeze-resistant and susceptible, winter wheat mutant lines. BMC Plant Biol 9:34–45. https://doi.org/10.1186/1471-2229-9-34
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinazachi K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathways under drought and high salinity conditions. Proc Natl Acad Sci USA 97:11632–11637. https://doi.org/10.1073/pnas.190309197
Wang J, Li Q, Mao X, Li A, Jing R (2016) Wheat Transcription Factor TaAREB3 Participates in Drought and Freezing Tolerances in Arabidopsis. Int J Biol Sci 12:257–269. https://doi.org/10.7150/ijbs.13538
Wei B, Jing R, Wang C, Chen J, Mao X, Chang X (2009) Dreb1, genes in wheat (triticum aestivum. L): development of functional markers and gene mapping based on SNPs. Mol Breed 23:13–22. https://doi.org/10.1007/s11032-008-9209-z
Yang Y, Al-Baidhani HHJ, Harris J, Riboni M, Li Y et al (2020) DREB/CBF expression in wheat and barley using the stress-inducible promoters of HD-Zip I genes: impact on plant development, stress tolerance and yield. Plant Biotechnol J 18:829–844. https://doi.org/10.1111/pbi.13252
Zotova L, Kurishbayev A, Jatayev S, Khassanova G, Zhubatkanov A et al (2018) Genes encoding transcription factors TaDREB5 and TaNFYC-A7 are differentially expressed in leaves of bread wheat in response to drought, dehydration and ABA. Front. Plant Sci 9:1441. https://doi.org/10.3389/fpls.2018.01441
Acknowledgements
The authors thank Dr. Ramesh Singh, Associate Professor, Department of plant pathology, SVP University of agriculture & technology Meerut, India, for all the necessary help to carry out this research. Thanks to Director research SVPUAT for providing all the necessary facilities to conduct the research smoothly.
Author information
Authors and Affiliations
Contributions
KS and SPS did the field and wet experiments and data analysis. KS write the manuscript. MKY supervised the work.
Corresponding authors
Ethics declarations
Conflict of interest
We solemnly declare that no conflict of interest to publish this manuscript.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Singh, K., Singh, S.P. & Yadav, M.K. Physio-biochemical assessment and CBF genes expression analysis in wheat under dehydration condition. Biologia 77, 1851–1860 (2022). https://doi.org/10.1007/s11756-022-01028-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11756-022-01028-4