Abstract
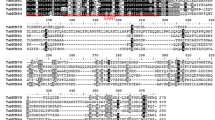
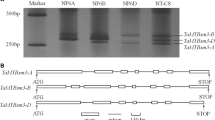
MYB proteins constitute one of the largest transcription factor families in plants and play critical roles in mediating plant adaptation to diverse abiotic stresses. In this study, fourty two of the MYB genes in wheat were subjected to the expression profile and function characterization analysis under phosphorus (Pi) deprivation. The results indicated that twenty-one, eleven, five, two, and one of the wheat MYB genes share homologous partners in B. Distachyon, rice, maize, barley, and Arabidopsis, respectively. Phylogenetic analysis suggested that the wheat MYB genes have evolved from five evolutionary pathways. Based on a microarray analysis, the wheat MYB genes were found to exhibit different transcripts under normal condition and can be grouped into various categories according to expression level, including those of high (eight members), moderate (ten members), low (ten members), and very low (fourteen members), respectively. Under Pi deprivation, most of the MYB genes showed significantly modified transcripts, in which TaMYB1, TaMYB16, and TaMYB73;1 were upregulated, and TaMYB11, TaMYB12, TaMYB13-2;1, and TaMYB;6 were downregulated. Transgenic analysis of TaMYB1, one of the upregulated genes, confirmed that it played an important role in mediating plant tolerance to Pi deprivation. The transgenic plants overexpressing TaMYB1 displayed higher plant drymass and more P accumulation than wild type under Pi deprivation. Together, our investigation indicates that a subset of wheat MYB genes involves plant response to Pi deficiency in which TaMYB1 plays critical roles in mediating plant tolerance to the Pi-starvation stress.




Similar content being viewed by others
References
Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15:63–78
Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK (2006) A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J Biol Chem 281:37636–37645
Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J (2010) A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet 6:e1001102
Chen B-J, Wang Y, Hu YL, Wu Q, Lin ZP (2005) Cloning and characteri zation of a drought inducible MYB gene from Boea crassifolia. Plant Sci 168:493–500
Cominelli E, Galbiati M, Vavasseur A, Conti L, Sala T, Vuylsteke M, Leonhardt N, Dellaporta SL, Tonelli C (2005) A guard cell-specific MYB transcription factor regulates stomatal movements and plant drought tolerance. Curr Biol 15:1196–1200
Cordell D, Drangert JO, White S (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19:292–305
Denekamp M, Smeekens SC (2003) Integration of wounding and osmotic stress signals determines the expression of the AtMYB102 transcription factor gene. Plant Physiol 132:1415–1423
Du H, Yang SS, Liang Z, Feng BR, Liu L, Huang YB, Tang YX (2012) Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol 12:106
Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L (2010) MYB transcription factors in Arabidopsis. Trends Plant Sci 15:573–581
El-kereamy A, Bi Y-M, Ranathunge K, Beatty PH, Good AG (2012) The rice R2R3-MYB transcription factor OsMYB55 is involved in the tolerance to high temperature and modulates amino acid metabolism. PLoS One 7(12):e52030
Gao J, Zhang Z, Peng R, Xiong A, Xu J, Zhu B, Yao QH (2010) Forced expression of MdMYB10, a myb transcription factor from apple, enhances tolerance to osmotic stress in transgenic Arabidopsis. Mol Biol Rep 38:205–211
Gigolashvili T, Yatusevich R, Berger B, Müller C, Flügge U (2007) The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 51:247–261
Glassop D, Smith SE, Smith FW (2005) Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222:688–698
Guo C, Zhao X, Liu X, Zhang L, Gu J, Li X, Lu W, Xiao K (2013) Function of wheat phosphate transporter gene TaPHT2;1 in Pi translocation and plant growth regulation under replete and limited Pi supply conditions. Planta 237:1163–1178
Guo C, Guo L, Li X, Gu J, Zhao M, Duan W, Ma C, Lu W, Xiao K (2014) TaPT2, a high-affinity phosphate transporter gene in wheat (Triticum aestivum L.), is crucial in plant Pi uptake under phosphorus deprivation. Acta Physiol Plant 36:1373–1384
Jin H, Martin C (1999) Multifunctionality and diversity within the plant MYB-gene family. Plant Mol Biol 41:577–585
Katiyar A, Smita S, Lenka SK, Rajwanshi R, Chinnusamy V, Bansal KC (2012) Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom 13:544
Kooiker M, Drenth J, Glassop D, McIntyre CL, Xue GP (2013) TaMYB13-1, a R2R3 MYB transcription factor, regulates the fructan synthetic pathway and contributes to enhanced fructan accumulation in bread wheat. J Exp Bot 64(12):3681–3696
Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW (2007) A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiol Plant 129:375–385
Li Q, Zhang C, Li J, Wang L, Ren Z (2012) Genome-wide identification and characterization of R2R3MYB family in Cucumis sativus. PLoS One 7:e47576
Li Z, Tian Y, Zhao W, Xu J, Wang L, Peng R, Yao Q (2015) Functional characterization of a grape heat stress transcription factor VvHsfA9 in transgenic Arabidopsis. Acta Physiol Plant 37:133
Liao Y, Zou HF, Wang HW, Zhang WK, Ma B, Zhang JS (2008) Soybean GmMYB76, GmMYB92, and GmMYB177 genes confer stress tolerance in transgenic Arabidopsis plants. Cell Res 18:1047–1060
Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K (2009) Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol 50:244–256
Martin C, Paz-Ares J (1997) MYB transcription factors in plants. Trends Genet 13:67–73
Rosinski JA, Atchley WR (1998) Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. J Mol Evol 46:74–83
Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J (2001) A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev 15:2122–2133
Schachtman DP, Reid RJ, Ayling SM (1998) Phosphorus uptake by plants: from soil to cell. Plant Physiol 116:447–453
Schünmann PHD, Richardson AE, Vickers CE, Delhaize E (2004) Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol 136:4205–4214
Seo PJ, Park CM (2010) MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol 186:471–483
Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM (2009) The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol 151:275–289
Seo PJ, Lee SB, Suh MC, Park MJ, Go YS, Park CM (2011) The MYB96 transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 23:1138–1152
Su CF, Wang YC, Hsieh TH, Lu CA, Tseng TH, Yu SM (2010) A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol 153:145–158
Su F, Zhang Q, Luo Z (2014) Isolation and characterisation of a Myb transcription factor DkPA1 related to proanthocyanidin biosynthesis in C-PCNA and non-PCNA persimmon (Diospyros kaki Thunb.) fruit. Acta Physiol Plant 36:1831–1839
Su L, Wang Y, Liu D, Li X, Zhai Y, Sun X, Li X, Liu Y, Li J, Wang Q (2015) The soybean gene, GmMYNJ2, eancodes a R2R3-type transcription factor involved in frought stress tolerance in Arabidopsis thaliana. Acta Physiol Plant 37:138
Sun Z, Ding C, Li X, Xiao K (2012) Molecular characterization and expression analysis of TaZFP15, a C2H2- type zinc finger transcription factor gene in wheat (Triticum aestivum L.). J Integr Agric 11:31–42
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conerved MYB recognition sequence. Plant Cell 5:1529–1539
Vance CP, Uhde-stone C, Allan DL (2003) Phosphorus acquisition and use: critical adaptations by plants for securing a non renewable resource. New Phytol 157:423–427
Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I (2004) Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J 37:115–127
Wang J, Sun J, Miao J, Guo J, Shi Z, He M, Chen Y, Zhao X, Li B, Han F, Tong Y, Li Z (2013) A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann Bot 111:1139–1153
Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM (2009) Expansion and diversification of the Populus R2R3-MYB family of transcription factors. Plant Physiol 149:981–993
Wu P, Xu J (2010) Does OsPHR2, central Pi-signaling regulator, regulate some unknown factors crucial for plant growth? Plant Signal Behav 5:712–714
Yang A, Dai X, Zhang WH (2012) A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J Exp Bot 63(7):2541–2556
Yanhui C, Xiaoyuan Y, Kun H, Meihua L, Jigang L, Zhaofeng G, Zhiqiang L, Yunfei Z, Xiaoxiao W, Xiaoming Q, Yunping S, Li Z, Xiaohui D, Jingchu L, Xing-Wang D, Zhangliang C, Hongya G, Li-Jia Q (2006) The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol Biol 60:107–124
Zhu J, Verslues PE, Zheng X, Lee B, Zhan X, Manabe Y, Sokolchik I, Zhu Y, Dong CH, Zhu JK, Hasegawa PM, Bressan RA (2005) Hos10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. PNAS 102:9966–9971
Acknowledgments
This work was supported by the National Natural Science Foundation of China (31371618 and 31201674), Natural Science Foundation of Hebei (C2015204057), and the Key Laboratory of Crop Growth Regulation of Hebei Province. The authors thank two anonymous reviewers and the editor whose detailed comments and careful work helped to improve the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by S. Abe.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Fang, W., Ding, W., Zhao, X. et al. Expression profile and function characterization of the MYB type transcription factor genes in wheat (Triticum aestivum L.) under phosphorus deprivation. Acta Physiol Plant 38, 5 (2016). https://doi.org/10.1007/s11738-015-1997-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1997-2