Abstract
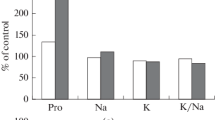
The morphological, biochemical and genetic characteristics of two Bassia sedoides (Chenopodiaceae) populations in the Southern Urals were studied. The plants of the Makan and Podolsk populations differ in growth (approximately 10-fold), in water and potassium contents and Na+/K+ ratio. On the basis of the genetic assay (by isozymes, RAPD and ISSR markers) of B. sedoides from the Makan and Podolsk populations, the intraspecific differences have been identified. The more productive Makan population showed greater genetic polymorphism, whereas the less productive Podolsk population showed less genetic polymorphism. The seedlings of B. sedoides from the Makan and Podolsk populations were cultivated under low and moderate salinity (100 and 200 mM NaCl, respectively) and equivalent osmoticity generated by the two PEG concentrations. Both populations were sensitive to dehydration initiated by PEG. Podolsk seedlings were more sensitive to osmotic stress and were characterised by an increase in proline content. Low salinity (100 mM NaCl) was necessary for optimal growth of seedlings from the Makan population. They showed significantly increased fresh biomass and number of lateral shoots. The maximal growth of seedlings from Podolsk was under 0–100 mM NaCl, and their biomass was approximately 1.4-fold lower than that of the Makan seedlings. Under moderate salinity (200 mM NaCl), the Makan seedlings were more salt tolerant than the Podolsk seedlings because of maintaining a low Na+/K+ ratio. Under natural conditions, the excess of Na+/K+ ratio compared with values for optimal growth under laboratory conditions was approximately threefold for the Makan plants and approximately fivefold for the Podolsk plants. High values of the Na+/K+ ratio under natural conditions indicate a deficit of potassium in the soil. Perhaps, the degree of potassium deficiency is one of the factors influencing the differences in productivity and the level of genetic variation of B. sedoides under natural conditions.

Similar content being viewed by others
References
Adams E, Shin R (2014) Transport, signalling, and homeostasis of potassium and sodium in plants. J Integr Plant Biol 56(3):231–249
Ashraf M, Harris PJC (2004) Potential biochemical indicator of salinity tolerance in plants. Plant Sci 166:3–16
Aslam R, Bostan N, Nabgha-e-Amen Maleeha Matia, Safdar W (2011) A critical review on halophytes: salt tolerant plants. J Med Plants Res 5:7108–7118
Bates LS, Waldren RP, Teare ID (1973) Rapid determination of free proline for water stress studies. Plant Soil 39:205–207
Bose J, Rodrigo-Moreno A, Lai D, Xie Y, Shen W, Shabala S (2015) Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann Bot 115:481–494
Chen Z, Pottosin II, Cuin TA, Fuglsang AT, Tester M, Jha D, Zepeda-Jazo I, Zhao M, Palmgren MG, Newman IA, Shabala S (2007) Root plasma membrane transporters controlling Na+/K+ homeostasis in salt-stressed barley. Plant Physiol 145:1714–1725
Flowers TJ, Colmer TD (2008) Salinity tolerance in halophytes. New Phytol 179:945–963
Flowers TJ, Colmer TD (2015) Plant salt tolerance: adaptation in halophytes. Ann Bot 155:327–331
Gunasekera CP, Martin LD, Siddique KHM, Walton GH (2006) Genotype by environment interactions of Indian mustard (Brassica juncea L.) and canola (B. napus L.) in Mediterranean-type environments. Crop growth and seed yield. Eur J Agr 25:1–12
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics 2014:701596. doi:10.1155/2014/701596
Hedrick PW (1985) Genetics of populations. Jones and Bartlett, Boston
Kadereit G, Freitag H (2011) Molecular phylogeny of Camphorosmeae (Camphorosmoideae, Chenopodiaceae): implications for biogeography, evolution of C4-photosynthesis and taxonomy. Taxon 60(1):51–78
Ma L, Zhang H, Sun L, Jiao Y, Zhang G, Miao Ch, Hao F (2012) NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J Exp Bot 63(1):305–317
Maathuis FJM (2014) Sodium in plants: perception, signaling, and regulation of sodium fluxes. J Exp Bot 3:849–858
Maathuis FJM, Antmann A (1999) K+ nutrition and Na+ toxicity: the basis of cellular K+/Na+ ratios. Ann Bot 84:123–133
Maathuis FJM, Ahmad I, Patishtan J (2014) Regulation of Na+ flues in plant. Front Plant Sci. doi:10.3389/fpls.2014.00467
Mandak B, Bimova K, Plackova I (2006) Genetic structure of experimental populations and reproductive fitness in a heterocarpic plant Atriplex tatarica (Chenopodiaceae). Am J Bot 93(11):1640–1649
Marden JH (2013) Nature’s inordinate fondness for metabolic enzymes: why metabolic enzyme loci are so frequently targets of selection. Mol Ecol 22:5743–5764
Megdiche W, Amor NB, Debez A, Hessini K, Ksuori R, Ya Zuily-Fodil, Abdelly Ch (2007) Salt tolerance of the annual halophyte Cakile maritima as affected by the provenance and the developmental stage. Acta Physiol Plant 29:375–384
Mitton JB, Grant MC, Yoshino AM (1998) Variation in allozymes and stomatal size in pinyon (Pinus edulis, Pinaceae), associated with soil moisture. Amer J Bot 85:1262–1265
Muona O, Szmidt AA (1985) Multilocus study of natural populations of Pinus sylvestris: population genetics in forestry. Lect Notes Biomath 60:226–240
Nei M, Roychoudhury AK (1974) Sampling variances of heterozygosity and genetic distance. Genetics 76:379–390
Nevo E, Krugman T, Beiles A (1994) Edaphic natural selection of allozyme polymorphisms in Aegilops peregrina at a Galilee microsite in Israel. Heredity 72:109–112
Sage RF, Sage TL, Kocacinarz F (2012) Photorespiration and the evolution of C4 photosynthesis. Annu Rev Biol 63:17.1–17.29
Shabala S, Pottosin II (2010) Potassium and potassium-permeable channels in plant salt tolerance. In: Demidchik V, Maathuis F (eds) Ion channels and plant stress responses, signaling and communication in plants. Springer, Heidelberg. doi:10.1007/978-3-642-10494-7_5
Shavrukov Yu (2013) Salt stress or salt shock: which genes are we studying? J Exp Bot 64(1):119–127
Soltis DE, Soltis PS (1990) Isozymes in plant biology. Springer, London
Spooner DR, van Treuren, de Vicente MC (2005) Molecular markers for genebank management. IPGRI Technical Bulletin No. 10. International Plant Genetic Resources Institute, Rome, Italy
Tipirdamaz R, Gagneul D, Duhaze´ C, Aı¨nouche A, Monnier C, o¨zkum D, Larher F (2005) Clustering of halophytes from an inland salt marsh in Turkey according to their ability to accumulate sodium and nitrogenous osmolytes. Env Exp Bot 57(1–2):139–153
Voznesenskaya EV, Koteyeva NK, Akhani H, Roalson EH, Edwards GE (2013) Structural and physiological analysis in Salsoleae (Chenopodiaceae) indicate multiple transitions among C3, intermediate, and C4 photosynthesis. J Exp Bot 64:3583–3604
Wang B, Davenport RJ, Volkov V, Amtmann A (2006) Low unidirectional sodium influx into root cells restricts net sodium accumulation in Thellungiella halophila, a salt-tolerant relative of Arabidopsis thaliana. J Exp Bot 57(5):1161–1170
Widodo Patterson JH, Newbigin E, Tester M, Bacic A, Roessner U (2009) Metabolic responses to salt stress of barley (Hordeum vulgare L.) cultivars, Sahara and Clipper, which differ in salinity tolerance. J Exp Bot 60:4089–4103
Wojnicka-Poltorak A, Chudzinska E, Shuiskay E, Barchak H, Toderich K, Prus-Glowacki W (2002) Izoenzymatic and cytolodical studies of some asiatic species of the genus Salsola. Acta Societatis Botanicorum Polonae 71(2):115–120
Yeh FC, Yang RC, Boyle T (1999) POPGEN, version 1.32. Microsoft Windows-based freeware for population genetic analysis. University of Alberta/CIFOR, Edmonton
Acknowledgments
This work was partially supported by the Russian Foundation for Basic Research (project no. 12_04_97023_r_a). The authors would like to thank Enago (www.enago.com) for the English language review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Apostol.
Rights and permissions
About this article
Cite this article
Shuyskaya, E., Rakhmankulova, Z., Voronin, P. et al. Salt and osmotic stress tolerances of the C3–C4 xero-halophyte Bassia sedoides from two populations differ in productivity and genetic polymorphism. Acta Physiol Plant 37, 236 (2015). https://doi.org/10.1007/s11738-015-1981-x
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1981-x