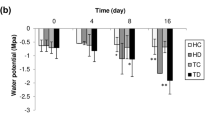
Abstract
A better understanding of drought response proteins may improve our understanding of the mechanisms underlying drought tolerance in sugarcane. In this research, drought-tolerant (K86-161) and drought-sensitive (B34-164) sugarcane cultivars were grown and exposed to drought stress. The changes in protein expression in leafs, leaf sheaths and roots were analyzed using proteomics techniques. Proteins that responded to drought in both cultivars could be classified into four major categories, including energy and metabolism, photosynthesis, antioxidant, and defense protein. Interestingly, an increased abundance of fructose-bisphosphate aldolase under drought was observed in all three organs of K86-161. Elevated expression of oxygen-evolving enhancer protein was also found in leaves and leaf sheaths of K86-161, when compared with their controls. Additionally, SOD was abundant in the leaves and roots of K86-161. Importantly, the expression level of these proteins decreased in B34-164 under drought stress. These contrasting results suggest that these proteins were inhibited by drought stress in the drought-sensitive cultivar. This proteomic research is the first to combine analyses of leaves, leaf sheaths and roots in sugarcane, which may enhance our understanding of drought responses at the molecular level and lead to selective breeding for enhanced drought tolerance.




Similar content being viewed by others
Abbreviations
- 2D-PAGE:
-
Two-dimensional polyacrylamide gel electrophoresis
- LC/MS-MS:
-
Liquid chromatography coupled to mass spectrometry
- FC:
-
Field capacity
- PWC:
-
Permanent wilting point
- IEF:
-
Isoelectric focusing
- PRK:
-
Phosphoribulokinase
- FNR:
-
Ferredoxin-NADP reductase
- RuBP:
-
d-Ribulose 1,5-bisphosphate
- ROS:
-
Reactive oxygen species
- SOD:
-
Superoxide dismutase
References
Abbasi FM, Komatsu S (2004) A proteomic approach to analyze salt-responsive proteins in rice leaf sheath. Proteomics 4:2072–2081
Ahsan N, Lee DG, Lee KW, Alam I, Lee SH, Bahk JD, Lee BH (2008) Glyphosate-induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol Biochem 46:1062–1070
Ail S, Munir A, Hayat R, Ijaz SS (2005) Enhancing water use efficiency, fixation capacity of mash bean and soil profile nitrate content with phosphorus and potassium application. Agron J 4:340–344
Ali GM, Komatsu S (2006) Proteomic analysis of rice leaf sheath during drought stress. J Proteome 5:396–403
Aranjuelo I, Molero G, Erice G, Avice JC, Nogues S (2011) Plant physiology and proteomics reveals the leaf response to drought in alfalfa (Medicagosativa L.). J Exp Biol 62:111–123
Azcarate-Peril MA, Bruno-Bárcena JM, Hassan HM, Klaenhammer TR (2006) Transcriptional and functional analysis of oxalyl-coenzyme A (CoA) decarboxylase and formyl-CoA transferase genes from Lactobacillus acidophilus. Appl Environ Microbiol 72:1891–1899
Balmer Y, Koller A, Val G, Schürmann Buchanan BB (2004) Proteomics uncovers proteins interacting electrostatically with thioredoxin in chloroplasts. Photosynth Res 79:275–280
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–302
Caruso G, Cavaliere C, Guarino C, Gubbiotti R, Foglia P, Lagana A (2008) Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal Bioanal Chem 391:381–390
Castillejo MA, Maldonado AM, Ogueta S, Jorrín JV (2008) Proteomic analysis of responses to drought stress in sunflower (Helianthus annuus) leaves by 2DE gel electrophoresis and mass spectrometry. The Open Proteomics J 1:59–71
Chaves MM, Maroco JP, Pereira J (2003) Understanding plant responses to drought-from genes to the whole plant. Functional Plant Biol 30:239–264
Chen F, Li Q, Sun L, He Z (2006) The rice14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res 13:53–64
Crafts-Brandner SJ, Salvucci ME (2000) Rubisco activase constrains the photosynthetic potential of leaves at high temperature and CO2. Plant Biol 97:13430–13435
Deeba F, Pandey AK, Ranjan S, Mishra M, Sharma YK, Shirke PA, Pandey V (2012) Physiological and proteomic respones of cotton (Gossypium herbaceum L.) to drought stress. Plant Physiol Biochem 53:6–18
Gazanchian A, Hajheidari M, Sima NK, Salekdeh GH (2007) Proteome response of Elymus elongatum to severe water stress and recovery. J Exp Bot 58:291–300
Ghannoum O (2009) C4 photosynthesis and water stress. Ann Bot 103:635–644
Hajheidari M, Abdollahian-Noghabi M, Askari H, Heidari M, Sadeghian SY, Ober ES, Salekdeh GH (2005) Proteome analysis of sugar beet leaves under drought stress. J Proteomics 5:950–960
Inman-Bamber NG (2004) Sugarcane water stress criteria for irrigation and drying off. Field Crop Res 89:107–122
Jangpromma N, Kitthaisong S, Komatsu S, Daduang S, Jaisil P, Thammasirirak S (2010a) A proteomics analysis of drought stress-responsive proteins as biomarker for drought-tolerant sugarcane cultivars. Am J Biochem Biotechnol 6:89–102
Jangpromma N, Songsri P, Thammasirirak S, Jaisil P (2010b) Rapid assessment of chlorophyll content in sugarcane using a SPAD chlorophyll meter across different water stress conditions. Asian J Plant Sci 9:368–374
Jangpromma N, Thammasirirak S, Jaisil P, Songsri P (2012) Effects of drought and recovery from drought stress on above ground and root growth, and water use efficiency in sugarcane (Saccharum officinarum L.). Aust J Crop Sci 6:1298–1304
Jangpromma N, Saito A, Araki T, Jaisil P, Songsri P, Songsri P, Daduang S, Kawaguchi Y, Dhiravisit A, Thammasirirak S (2014) Molecular cloning and characterization in eukaryotic expression systems of a sugarcane cysteine protease inhibitor gene involved in drought tolerance. Tur J Bot 38:724–736
Katam R, Basha SM, Vasanthaiah HN, Naik N (2007) Identification of drought tolerant groundnut genotypes employing proteomics approach. Agric Res 5:1–4
Klubicova K, Danchenko M, Miernyk JA, Rashydov NM, Berezhna VV, Ova AP, Hajduch M (2010) Proteomics analysis of flax grown in chernobyl area suggests limited effect of contaminated environment on seed proteome. Environ Sci Technol 44:6940–6946
Konishi H, Yamane H, Maeshima M, Komatsu S (2005) Characterization of fructose-bisphosphate aldolase regulated by gibberellin in roots of rice seedling. Plant Mol Biol 56:839–848
Lee J, Seo KJ (2004) Post-translational modifications and their biological functions: proteomic analysis and systematic approaches. J Biochem Mol Biol 37:35–44
Lee DG, Ahsan N, Lee SH, Lee JJ, Bahk JD, Kang KY, Lee BH (2009) Chilling stress-induced proteomic changes in rice roots. J Plant Physiol 166:1–11
Li K, Xu C, Zhang K, Yang A, Zhang J (2007) Proteomic analysis of roots growth and metabolic changes under phosphorus deficit in maize (Zea mays L.) plants. J Proteomics 7:1501–1512
Lu W, Tang X, Huo X, Xu R, Qi S, Huang J, Zheng C, Wu C (2012) Identification and characterization of fructose 1,6-bisphosphate aldolase genes in Arabidopsis reveal gene family with diverse responses to abiotic stresses. Gene 503:65–74
Merewitz EB, Gianfagna T, Huang B (2011) Protein accumulation in leaves and roots associated with improved drought tolerance in creeping bentgrass expressing an ipt gene for cytokinin synthesis. J Exp Bot 62:5311–5333
Miller DM (1985) Studies of root function in zea mays: iii. Xylem sap composition at maximum root pressure provides evidence of active transport into the xylem and a measurement of the reflection coefficient of the root. Plant Physiol Biochem 77:162–167
Mitra J (2001) Genetics and genetic improvement of drought resistance in crop plants. Curr Sci 80:758–763
Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI (2007) Photoinhibition of photosystem II under environmental stress. Biochim Biophys Acta 1767:414–421
O’ Farrell PH (1975) High resolution two-dimensional electrophoresis of protein. Biol Chem 250:4007–4021
Panter S, Thomson R, Bruxelles Gd, Laver D, Trevaskis B, Udvardi M (1999) Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Am Phytopath Soc 13:325–333
Pradet A, Raymond P (1983) Adenine nucleotide ratios and adenylate energy charge in energy metabolism. J Plant Physiol 34:199–200
Prajanban BO, Shawsuan L, Daduang S, Kommanee J, Roytrakul S, Dhiravisit A, Thammasirirak S (2012) Identification of five reptile egg whites protein using MALDI-TOF mass spectrometry and LC/MS-MS analysis. J Proteomics 75:1940–1959
Qaisar U, Irfan M, Meqbool A, Zahoor M, Khan MY, Rashid B, Riazuddin S, Husnain T (2009) Identification, sequencing and characterization of a stress induced homologue of fructose bisphosphate aldolase from cotton. Can J Plant Sci 90:41–48
Rabello AR, Guimarães CM, Rangel PHN, da Silva FR, Seixas D, de Souza E, Brasileiro ACM, Spehar CR, Ferreira ME, Mehta A (2008) Identification of drought-responsive genes in roots of upland rice (Oryza sativa L). BMC Genom 9:485
Riccardi F, Gazeau P, Vienne D, Zivy M (1998) Protein changes in response to progressive water deficit in maize. Plant Physiol 117:1253–1263
Ripley BS, Gilbert ME, Ibrahim DG, Osborne CP (2007) Drought constraints on C4 photosynthesis: stomatal and metabolic limitations in C3 and C4 subspecies of Alloteropsis semialata. J Exp Mar Biol Ecol 58:1351–1363
Ruiz-Lozano JM, Azcon R, Gòmez M (1996) Alleviation of salt stress by arbuscular-mycorrhizal Glomus species in Lactuca sativa plants. Physiol Plant 98:767–772
Salekdeh GH, Siopongco J, Wade LJ, Ghareyazie B, Bennett J (2002) Proteomic analysis of rice leaves during drought stress and recovery. J Proteomics 2:1131–1145
Shen S, Jing Y, Kuang T (2003) Proteomics approach to identify wound-response related proteins from rice leaf sheath. J Proteomics 3:527–535
Smit MA, Singels A (2006) The response of sugarcane canopy development to water stress. Field Crop Res 98:91–97
Songsri P, Vorasoot N, Jogloy S, Kesmala T, Akkasaeng C, Patanothai A, Holbrook CC (2009) Evaluation of yield and reproductive efficiency in peanut (Arachis hypogaea L.) under different available soil water. Asian J Plant Sci 8:465–473
Sugiharto B (2004) Biochemical and molecular studies on sucrose-phosphate synthase and drought inducible protein in sugarcane (Saccharum offcinarum). J Ilmu Dasar 5:62–67
Sugiharto B, Ernawati N, Mori H, Aoki K, Yonekura-Sakakibara K, Yamaya T, Sugiyama T, Sakahibara H (2002) Identification and characterization of a gene encoding drought-inducible protein localization in the binder sheath cell of sugarcane. Plant Cell Physiol 43:350–354
Tezara W, Mitchell VJ, Driscoll SD, Lawlor DW (1999) Water stress inhibits plant photosynthesis by decreasing coupling factor and ATP. Letters to Nature 401:114–117
Xiao X, Yanga F, Zhanga S, Korpelainenc H, Li C (2009) Physiological and proteomic responses of two contrasting Populus cathayana populations to drought stress. Physiol Plant 136:150–168
Xu WF, Shi WM (2006) Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: analysis by real-time RT-PCR. Ann Bot 98:965–974
Yan S, Tang Z, Su W, Sun W (2005) Proteomic analysis of salt stress-responsive proteins in rice root J Proteomics 5:235–244
Yan SP, Zhang QY, Tang ZC, Su WA, Sun WN (2006) Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol Cell Proteomics 5:484–496
Yoshimura K, Masuda A, Kuwano M, Yokota A, Akashi K (2008) Programmed proteome response for drought avoidance/tolerance in the root of a C3 xerophyte (wild Watermelon) under water deficits. Plant Cell Physiol 49:226–242
Zang X, Komatsu S (2007) A proteomics approach for identifying osmotic-stress-related proteins in rice. Phytochemistry 68:426–437
Zhaoa Y, Dua H, Wanga Z, Huanga B (2011) Identification of proteins associated with water-deficit tolerance in C4 perennial grass species, Cynodondactylon × Cynodon transvaalensis and Cynodon dactylon. Physiol Plant 141:40–55
Acknowledgments
This study was financially supported by the National Research University Project through the Bio-Fuel Research Cluster and Protein and Proteomics Research Center for Commercial and Industrial Purposes (ProCCI), Department of Biochemistry, Faculty of Science, Khon Kaen University. This study was supported (in part) by Northeast Thailand Cane and Sugar Research Center Khon Kaen University. We are most grateful to Mr. Werapon Ponragdee, Khon Kaen Field Crop Research Center, for providing sugarcane cultivars.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Hajduch.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11738_2015_1826_MOESM1_ESM.tif
Supplementary Fig. 1 Experimental design; the sixteen trees used for the experiments were randomly sampled from a heterogeneous population. The greenhouse conditions were used for growing the 2 sugarcane cultivars, K86-161 and B34-164 randomized by technical stages of stress to avoid instrumental drift and differences between batches. The white square indicates the well-watered control and gray squares indicate the drought-stressed plants for sugarcane K86-161 cultivar, respectively. The white triangles indicate well-watered (TIFF 3253 kb)
11738_2015_1826_MOESM2_ESM.tif
Supplementary Fig. 2 The replication of K86-161 and B34-164 sugarcane cultivars 2D gel. (a) and (b) show 2D- in leaf from well-watered control and drought-stressed K86-161 cultivar, respectively. (c) and (d) show 2D- in leaf from well-watered control and drought-stressed B34-164 cultivar, respectively. (e) and (f) show 2D- in leaf sheath from well-watered control and drought-stressed K86-161 cultivar, respectively. (g) and (h) show 2D- in leaf sheath from well-watered control and drought-stressed B34-164 cultivar, respectively. (i) and (j) show 2D- in root from well-watered control and drought-stressed K86-161cultivar, respectively. (k) and (l) show 2D- in root from well-watered control and drought-stressed B34-164 cultivar, respectively (TIFF 16309 kb)
11738_2015_1826_MOESM3_ESM.docx
Supplementary Table 1. Information about matched peptide sequence, changes in expression and organism sources of drought-responsive protein in leaves, leaf sheaths and root of the K86-161 and B34-164 cultivars that were identified by LC/MS–MS (DOCX 482 kb)
11738_2015_1826_MOESM4_ESM.xlsx
Supplementary Table 2 The relative volumes of all protein spots responding to drought in leaves, leaf sheaths and root of the K86-161 and B34-164 cultivars (XLSX 18 kb)
Rights and permissions
About this article
Cite this article
Khueychai, S., Jangpromma, N., Daduang, S. et al. Comparative proteomic analysis of leaves, leaf sheaths, and roots of drought-contrasting sugarcane cultivars in response to drought stress. Acta Physiol Plant 37, 88 (2015). https://doi.org/10.1007/s11738-015-1826-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11738-015-1826-7