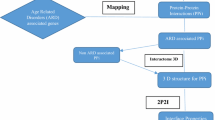
Abstract
Interactions between proteins largely govern cellular processes and this has led to numerous efforts culminating in enormous information related to the proteins, their interactions and the function which is determined by their interactions. The main concern of the present study is to present interface analysis of cardiovascular-disorder (CVD) related proteins to shed lights on details of interactions and to emphasize the importance of using structures in network studies. This study combines the network-centred approach with three dimensional studies to comprehend the fundamentals of biology. Interface properties were used as descriptors to classify the CVD associated proteins and non-CVD associated proteins. Machine learning algorithm was used to generate a classifier based on the training set which was then used to predict potential CVD related proteins from a set of polymorphic proteins which are not known to be involved in any disease. Among several classifying algorithms applied to generate models, best performance was achieved using Random Forest with an accuracy of 69.5 %. The tool named CARDIO-PRED, based on the prediction model is present at http://www.genomeinformatics.dce.edu/CARDIO-PRED/. The predicted CVD related proteins may not be the causing factor of particular disease but can be involved in pathways and reactions yet unknown to us thus permitting a more rational analysis of disease mechanism. Study of their interactions with other proteins can significantly improve our understanding of the molecular mechanism of diseases.





Similar content being viewed by others
References
Adie EA, Adams RR, Evans KL, Porteous DJ, Pickard BS (2006) SUSPECTS: enabling fast and effective prioritization of positional candidates. Bioinformatics 22(6):773–774
Aung Z (2006) Computational analysis of 3D protein structures. Doctoral dissertation, School of Computing, National Institute of Singapore
Basse MJ, Betzi S, Bourgeas R, Bouzidi S, Chetrit B, Hamon V, Morelli X, Roche P (2013) 2P2Idb: a structural database dedicated to orthosteric modulation of protein–protein interactions. Nucleic Acids Res 41:D824–D827
Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK (2006) Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet 38(5):576–582. doi:10.1038/ng1776
Choura M, Rebaï A (2011) Structural analysis of hubs in human NR-RTK network. Biol Direct 6:49. doi:10.1186/1745-6150-6-49
Chuang HY, Lee E, Liu YT, Lee D, Ideker T (2007) Network-based classification of breast cancer metastasis. Mol Syst Biol 3:140
Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabási AL (2007) The human disease network. Proc Natl Acad Sci 104(21):8685–8690
Gonzalez MW, Kann MG (2012) Protein interactions and disease. PLoS Comput Biol 8(12):e1002819. doi:10.1371/journal.pcbi.1002819
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software: an update SIGKDD explorations 11(1)
Kann MG (2007) Protein interactions and disease: computational approaches to uncover the etiology of diseases. Brief Bioinform 8(5):333–346
Kar G, Gursoy A, Keskin O (2009) Human cancer protein–protein interaction network: a structural perspective. PLoS Comput Biol. doi:10.1371/journal.pcbi.1000601
López-Bigas N, Ouzounis CA (2004) Genome-wide identification of genes likely to be involved in human genetic disease. Nucl Acids Res 32(10):3108–3114
Magrane M, The UniProt consortium (2011) UniProt knowledgebase: a hub of integrated protein data
Marini F, Brandi ML (2010) Genetic determinants of osteoporosis: common bases to cardiovascular diseases. Int J Hypertens. doi:10.4061/2010/394579
Moreira IS, Fernandes PA, Ramos MJ (2007) Hot spots—a review of the protein–protein interface determinant amino-acid residues. Proteins 68(4):803–812
Mosca R, Céol A, Aloy P (2013) Interactome3D: adding structural details to protein networks. Nat Methods 10(1):47–53
North BJ, Sinclair DA (2012) The intersection between aging and cardiovascular disease. Circ Res 110:1097–1108
Online Mendelian Inheritance in Man, OMIM®. McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University (Baltimore, MD). http://www.omim.org/. Accessed April 2014
Rao A, Bulusu G, Srinivasan R, Joseph T (2012) Protein–protein interactions and disease, protein interactions. Dr. Jianfeng Cai (Ed.), ISBN: 978-953-51-0244-1, InTech
Wang X, Wei X, Thijssen B, Das J, Lipkin SM, Yu H (2012) Three-dimensional reconstruction of protein networks provides insight into human genetic disease. Nat Biotechnol 30(2):159–164. doi:10.1038/nbt.2106
Zhang KX, Ouellette BF (2011) CAERUS: predicting CAncER oUtcomeS using relationship between protein structural information, protein networks, gene expression data, and mutation data. PLoS Comput Biol. doi:10.1371/journal.pcbi.1001114
Acknowledgments
The study has been supported by SERB under OYS scheme via Project File No. SR/FT/LS-96/2011.
Conflict of interest
The author(s) declare that they have no competing interests.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jain, P., Thukral, N., Gahlot, L.K. et al. CARDIO-PRED: an in silico tool for predicting cardiovascular-disorder associated proteins. Syst Synth Biol 9, 55–66 (2015). https://doi.org/10.1007/s11693-015-9164-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11693-015-9164-z