Abstract
Purpose
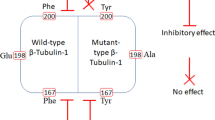
Fasciola hepatica is a globally distributed trematode that causes significant economic losses. Triclabendazole is the primary pharmacological treatment for this parasite. However, the increasing resistance to triclabendazole limits its efficacy. Previous pharmacodynamics studies suggested that triclabendazole acts by interacting mainly with the β monomer of tubulin.
Methods
We used a high-quality method to model the six isotypes of F. hepatica β-tubulin in the absence of three-dimensional structures. Molecular dockings were conducted to evaluate the destabilization regions in the molecule against the ligands triclabendazole, triclabendazole sulphoxide and triclabendazole sulphone.
Results
The nucleotide binding site demonstrates higher affinity than the binding sites of colchicine, albendazole, the T7 loop and pβVII (p < 0.05). We suggest that the binding of the ligands to the polymerization site of β-tubulin can lead a microtubule disruption. Furthermore, we found that triclabendazole sulphone exhibited significantly higher binding affinity than other ligands (p < 0.05) across all isotypes of β-tubulin.
Conclusions
Our investigation has yielded new insight on the mechanism of action of triclabendazole and its sulphometabolites on F. hepatica β-tubulin through computational tools. These findings have significant implications for ongoing scientific research ongoing towards the discovery of novel therapeutics to treat F. hepatica infections.





Similar content being viewed by others
Data Availability
The data that generated this publication is available if required by this journal. All the data is freely accessible online, the models are in the repository of protein models reported in the results.
References
Mas-Coma S, Valero MA, Bargues MD (2009) Chapter 2. Fasciola, lymnaeids and human fascioliasis, with a global overview on disease transmission, epidemiology, evolutionary genetics, molecular epidemiology and control. Adv Parasitol 69:41–146. https://doi.org/10.1016/S0065-308X(09)69002-3
Mccann C, Baylis M, Williams D (2010) The development of linear regression models using environmental variables to explain the spatial distribution of Fasciola hepatica infection in dairy herds in England and Wales. Int J Parasitol 40:1021–1028. https://doi.org/10.1016/j.ijpara.2010.02.009
Espinoza JR, Terashima A, Herrera-Velit P, Marcos LA (2010) Fasciolosis Humana y Animal en el Perú: Impacto en la Economía de las Zonas Endémicas. Rev Peru Med Exp Salud Publica 4:604–612
Schweizer G, Braun U, Deplazes P, Torgerson PR (2005) Estimating the financial losses due to bovine fasciolosis in Switzerland. Vet Rec 157:188–193. https://doi.org/10.1136/vr.157.7.188
Charlier J, Vercruysse J, Morgan E et al (2014) Recent advances in the diagnosis, impact on production and prediction of Fasciola hepatica in cattle. Parasitology 141:326–335. https://doi.org/10.1017/S0031182013001662
World Health Organization (WHO) (2013) Sustaining the drive to overcome the global impact of neglected tropical diseases: second WHO report on neglected tropical diseases. Geneva xii; 140. ISBN:9789241564540
Fairweather I, Boray JC (1999) Fasciolicides efficacy, actions, resistance and its management. Vet J 158:81–112
Hennessy DR, Lacey E, Steel JW, Prichard RK (1987) The kinetics of triclabendazole disposition in sheep. J Vet Pharmacol Ther 10:64–72. https://doi.org/10.1111/j.1365-2885.1987.tb00078.x
Barrera B, Otero JA, Egido E et al (2012) The anthelmintic triclabendazole and its metabolites inhibit the membrane transporter ABCG2/BCRP. Antimicrob Agents Chemother 56:3535–3543. https://doi.org/10.1128/AAC.06345-11
Brennan GP, Fairweather I, Trudgett A et al (2007) Understanding triclabendazole resistance. Exp Mol Pathol 82:104–109. https://doi.org/10.1016/j.yexmp.2007.01.009
Abdelaal MMO, Brennan GP, Abdel-Aziz A, Fairweather I (2017) Ultrastructural changes to the tegumental system and gastrodermal cells of adult Fasciola hepatica following treatment in vivo with a commercial preparation of myrrh (Mirazid). J Helminthol 91:672–685. https://doi.org/10.1017/S0022149X16000705
Fairweather I (2005) Triclabendazole: new skills to unravel an old(ish) enigma. J Helminthol 79:227–234. https://doi.org/10.1079/joh2005298
Fairweather I (2009) Triclabendazole progress report, 2005–2009: an advancement of learning? J Helminthol 83:139–150. https://doi.org/10.1017/S0022149X09321173
Fairweather I (2011) Reducing the future threat from (liver) fluke: realistic prospect or quixotic fantasy? Vet Parasitol 180:133–143. https://doi.org/10.1016/j.vetpar.2011.05.034
McConville M, Brennan GP, McCoy M et al (2007) Immature triclabendazole-resistant Fasciola hepatica: tegumental responses to in vitro treatment with the sulphoxide metabolite of the experimental fasciolicide compound alpha. Parasitol Res 100:365–377. https://doi.org/10.1007/s00436-006-0270-4
Hanna R (2015) Fasciola hepatica: histology of the reproductive organs and differential effects of triclabendazole on drug-sensitive and drug-resistant fluke isolates and on flukes from selected field cases. Pathogens 4:431–456. https://doi.org/10.3390/pathogens4030431
Abdelaal MMO, Brennan GP, Hanna REB et al (2017) Disruption of egg production by triclabendazole-resistant Fasciola hepatica following treatment with a commercial preparation of myrrh (Mirazid). Acta Parasitol 62:336–347. https://doi.org/10.1515/ap-2017-0041
Stitt AW, Fairweather I (1996) Fasciola hepatica: disruption of the vitelline cells in vitro by the sulphoxide metabolite of triclabendazole. Parasitol Res 82:333–339. https://doi.org/10.1007/s004360050122
McConville M, Brennan GP, McCoy M et al (2006) Adult triclabendazole-resistant Fasciola hepatica: surface and subsurface tegumental responses to in vitro treatment with the sulphoxide metabolite of the experimental fasciolicide compound alpha. Parasitology 133:195–208. https://doi.org/10.1017/S0031182006000114
Robinson MW, Trudgett A, Hoey EM, Fairweather I (2002) Triclabendazole-resistant Fasciola hepatica: β-tubulin and response to in vitro treatment with triclabendazole. Parasitology 124:325–338. https://doi.org/10.1017/S003118200100124X
Halferty L, Brennan GP, Trudgett A et al (2009) Relative activity of triclabendazole metabolites against the liver fluke, Fasciola hepatica. Vet Parasitol 159:126–138. https://doi.org/10.1016/j.vetpar.2008.10.007
Nogales E (2015) An electron microscopy journey in the study of microtubule structure and dynamics. Protein Sci 24:1912–1919. https://doi.org/10.1002/pro.2808
Nogales E, Wolf SG, Downing KH (1998) Structure of the ab tubulin dimer by electron crystallography. Nature 8:199–203. https://doi.org/10.1107/S2052252519011497
Aguayo-Ortiz R, Méndez-Lucio O, Medina-Franco JL et al (2013) Towards the identification of the binding site of benzimidazoles to β-tubulin of Trichinella spiralis: insights from computational and experimental data. J Mol Graph Model 41:12–19. https://doi.org/10.1016/j.jmgm.2013.01.007
Aguayo-Ortiz R, Méndez-Lucio O, Romo-Mancillas A et al (2013) Molecular basis for benzimidazole resistance from a novel β-tubulin binding site model. J Mol Graph Model 45:26–37. https://doi.org/10.1016/j.jmgm.2013.07.008
Ryan LA, Hoey E, Trudgett A et al (2008) Fasciola hepatica expresses multiple α- and β-tubulin isotypes. Mol Biochem Parasitol 159:73–78. https://doi.org/10.1016/j.molbiopara.2008.02.001
Robinson MW, McFerran N, Trudgett A et al (2004) A possible model of benzimidazole binding to β-tubulin disclosed by invoking an inter-domain movement. J Mol Graph Model 23:275–284. https://doi.org/10.1016/j.jmgm.2004.08.001
Chambers E, Ryan LA, Hoey EM et al (2010) Liver fluke β-tubulin isotype 2 binds albendazole and is thus a probable target of this drug. Parasitol Res 107:1257–1264. https://doi.org/10.1007/s00436-010-1997-5
Mühlethaler T, Gioia D, Prota AE et al (2021) Comprehensive analysis of binding sites in tubulin. Angew Chem Int Ed 60:13331–13342. https://doi.org/10.1002/anie.202100273
Naaz F, Haider MR, Shafi S, Yar MS (2019) Anti-tubulin agents of natural origin: targeting taxol, vinca, and colchicine binding domains. Eur J Med Chem 171:310–331. https://doi.org/10.1016/j.ejmech.2019.03.025
Khrapunovich-Baine M, Menon V, Yang CPH et al (2011) Hallmarks of molecular action of microtubule stabilizing agents: effects of epothilone B, ixabepilone, peloruside A, and laulimalide on microtubule conformation. J Biol Chem 286:11765–11778. https://doi.org/10.1074/jbc.M110.162214
Berman HM, Battistuz T, Bhat TN et al (2000) The protein data bank. Nucleic Acids Res 8:235–242. https://doi.org/10.1107/S0907444902003451
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Nogales E, Downing KH, Amos LA, Lowe J (1998) Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol 5:451–458
Salentin S, Schreiber S, Haupt VJ et al (2015) PLIP: fully automated protein-ligand interaction profiler. Nucleic Acids Res 43:W443–W447. https://doi.org/10.1093/nar/gkv315
Jumper J, Evans R, Pritzel A et al (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596:583–589. https://doi.org/10.1038/s41586-021-03819-2
Mirdita M, Schütze K, Moriwaki Y et al (2022) ColabFold: making protein folding accessible to all. Nat Methods 19:679–682. https://doi.org/10.1038/s41592-022-01488-1
Hornak V, Abel R, Okur A et al (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins Struct Funct Genet 65:712–725. https://doi.org/10.1002/prot.21123
Guex N, Peitsch MC, Schwede T (2009) Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: a historical perspective. Electrophoresis 30:162–173. https://doi.org/10.1002/elps.200900140
Chen VB, Arendall WB, Headd JJ et al (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr 66:12–21. https://doi.org/10.1107/S0907444909042073
Waterhouse A, Bertoni M, Bienert S et al (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296–W303. https://doi.org/10.1093/nar/gky427
Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 35:407–410. https://doi.org/10.1093/nar/gkm290
Sippl MJ (1993) Recognition of errors in three-dimensional structures of proteins. Proteins Struct Funct Bioinform 17:355–362. https://doi.org/10.1002/prot.340170404
Abraham MJ, Murtola T, Schulz R et al (2015) Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2:19–25. https://doi.org/10.1016/j.softx.2015.06.001
Huang J, Rauscher S, Nawrocki G et al (2016) CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Methods 14:71–73. https://doi.org/10.1038/nmeth.4067
Maxwell JC (1890) The scientific papers. Cambridge University Press, Cambridge. https://doi.org/10.1017/CBO9780511710377
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190. https://doi.org/10.1063/1.328693
Kim S, Thiessen PA, Bolton EE et al (2016) PubChem substance and compound databases. Nucleic Acids Res 44:D1202–D1213. https://doi.org/10.1093/nar/gkv951
Trott O, Olson AJ (2010) Software news and update AutoDock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461
Dallakyan S, Olson AJ (2015) Small-molecule library screening by docking with PyRx. Methods Mol Biol 1263:243–250. https://doi.org/10.1007/978-1-4939-2269-7_19
Halgren TA (1996) Merck molecular force field. V. Extension of MMFF94 using experimental data, additional computational data, and empirical rules. J Comput Chem 17:616–641. https://doi.org/10.1002/(SICI)1096-987X(199604)17:5/6%3c616::AID-JCC5%3e3.0.CO;2-X
Löwe J, Li H, Downing KH, Nogales E (2001) Refined structure of αβ-tubulin at 3.5 Å resolution. J Mol Biol 313:1045–1057. https://doi.org/10.1006/jmbi.2001.5077
Castrignanò T, De Meo PD, Cozzetto D et al (2006) The PMDB protein model database. Nucleic Acids Res 34:D306–D309. https://doi.org/10.1093/nar/gkj105
Chemale G, Perally S, Lacourse EJ et al (2010) Comparative proteomic analysis of triclabendazole response in the liver fluke Fasciola hepatica. J Proteome Res 9:4940–4951. https://doi.org/10.1021/pr1000785
Radio S, Fontenla S, Solana V et al (2018) Pleiotropic alterations in gene expression in Latin American Fasciola hepatica isolates with different susceptibility to drugs. Parasit Vectors 11:1–11. https://doi.org/10.1186/s13071-017-2553-2
Desai A, Mitchison TJ (1997) Microtubule polymerization dynamics. Annu Rev Cell Dev Biol 13:83–117. https://doi.org/10.1146/annurev.cellbio.13.1.83
Amos LA, Lowe J (1999) How Taxol(R) stabilises microtubule structure. Chem Biol 6:R65–R69
Von Samson-Himmelstjerna G, Blackhall WJ, McCarthy JS, Skuce PJ (2007) Single nucleotide polymorphism (SNP) markers for benzimidazole resistance in veterinary nematodes. Parasitology 134:1077–1086. https://doi.org/10.1017/S0031182007000054
Kwa MS, Veenstra JG, Roos MH (1994) Benzimidazole resistance in Haemonchus contortus is correlated with a conserved mutation at amino acid 200 in beta-tubulin isotype 1. Mol Biochem Parasitol 63:299–303. https://doi.org/10.1016/0166-6851(94)90066-3
Fuchs MA, Ryan LA, Chambers EL et al (2013) Differential expression of liver fluke β-tubulin isotypes at selected life cycle stages. Int J Parasitol 43:1133–1139. https://doi.org/10.1016/j.ijpara.2013.08.007
Lipkowitz KB, McCracken RO (1991) A molecular modeling approach to in vivo efficacy of triclabendazole. J Parasitol 77:998–1005. https://doi.org/10.2307/3282756
Fairweather I, Brennan GP, Robinson MW, Skuce PJ (2020) Drug resistance in liver flukes. Int J Parasitol Drugs Drug Resist. https://doi.org/10.1016/j.ijpddr.2019.11.003
Lubega GW, Prichard RK (1991) Interaction of benzimidazole anthelmintics with Haemonchus contortus tubulin: binding affinity and anthelmintic efficacy. Exp Parasitol 73:203–213. https://doi.org/10.1016/0014-4894(91)90023-P
Jiménez-González A, De Armas-Serra C, Criado-Fornelio A et al (1991) Preliminary characterization and interaction of tubulin from Trichinella spiralis larvae with benzimidazole derivatives. Vet Parasitol 39:89–99. https://doi.org/10.1016/0304-4017(91)90065-4
Lu Y, Chen J, Xiao M et al (2012) An overview of tubulin inhibitors that interact with the colchicine binding site. Pharm Res 29:2943–2971. https://doi.org/10.1007/s11095-012-0828-z
Duan Y, Liu W, Tian L et al (2019) Targeting tubulin-colchicine site for cancer therapy: inhibitors, antibody–drug conjugates and degradation agents. Curr Top Med Chem 19:1289–1304. https://doi.org/10.2174/1568026619666190618130008
Barbier P, Dorléans A, Devred F et al (2010) Stathmin and interfacial microtubule inhibitors recognize a naturally curved conformation of tubulin dimers. J Biol Chem 285:31672–31681. https://doi.org/10.1074/jbc.M110.141929
Ravelli RBG, Gigant B, Curmi PA et al (2004) Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature 428:198–202. https://doi.org/10.1038/nature02393
Fetterer RH (1986) The effect of albendazole and triclabendazole on colchicine binding in the liver fluke Fasciola hepatica. J Vet Pharmacol Ther 9:49–54. https://doi.org/10.1111/j.1365-2885.1986.tb00011.x
Moudi M, Go R, Yien CYS, Nazre M (2013) Vinca alkaloids. Int J Prev Med 4:1231–1235
Zhou XJ, Rahmani R (1992) Preclinical and clinical pharmacology of vinca alkaloids. Drugs 44(Suppl 4):1–9. https://doi.org/10.2165/00003495-199200444-00002
Gigant B, Wang C, Ravelli RBG et al (2005) Structural basis for the regulation of tubulin by vinblastine. Nature 435:519–522. https://doi.org/10.1038/nature03566
Wang C, Cormier A, Gigant B, Knossow M (2007) Insight into the GTPase activity of tubulin from complexes with stathmin-like domains. Biochemistry 46:10595–10602. https://doi.org/10.1021/bi701147f
Mottier L, Virkel G, Solana H et al (2004) Triclabendazole biotransformation and comparative diffusion of the parent drug and its oxidized metabolites into Fasciola hepatica. Xenobiotica 34:1043–1057. https://doi.org/10.1080/00498250400015285
Acknowledgements
P.O.F. is grateful for the support of ANID-PCHA/2017/21170159.
Funding
This study did not receive funding from private sources.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by PO-F and JFB. The first draft of the manuscript was written by PO-F and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethics Approval
This study did not involve human or animal experimentation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Olivares-Ferretti, P., Beltrán, J.F., Salazar, L.A. et al. Protein Modelling and Molecular Docking Analysis of Fasciola hepatica β-Tubulin’s Interaction Sites, with Triclabendazole, Triclabendazole Sulphoxide and Triclabendazole Sulphone. Acta Parasit. 68, 535–547 (2023). https://doi.org/10.1007/s11686-023-00692-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-023-00692-z