Abstract
Purpose
A 2016 study estimated that over 3 billion people are currently at risk of contracting malaria. Although a wide variety of medications are available to treat malaria, the parasites have started to exhibit resistance to many commonly used therapeutics necessitating a push for new investigations to identify novel drug targets.
Methods
In this study, nucleic acid-binding Alba superfamily proteins of the human malaria parasite, Plasmodium falciparum were investigated to identify interacting protein motifs. A high-throughput molecular screening technique, phage display, coupled with next-generation sequencing was applied to assess large data sets.
Results
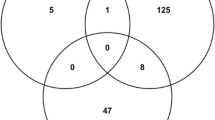
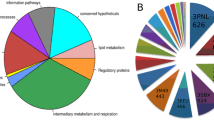
Four P. falciparum Alba proteins were used for screening which appear to have distinct roles in parasite biology based on the results of this work. The majority of the peptide motifs identified from phage display were involved in post-translational modification pathways, thus suggesting that parasite-specific gene regulatory mechanisms are involved which could serve as drug targets for novel therapeutics.
Conclusion
This study found 18 peptide motifs which potentially have strong interactions with one or more of the Alba superfamily proteins from P. falciparum. Considering the large fraction of post-translational modification-related peptide motifs identified from this work, one or more of the protein modification pathways could serve as a good target for malaria treatment.

Similar content being viewed by others
Availability of data and materials
The datasets generated during the current study are available from the corresponding author upon reasonable request.
References
WHO (2017) World Malaria Report 2017. 2017
Achan J et al (2011) Quinine, an old anti-malarial drug in a modern world: role in the treatment of malaria. Malar J 10:144. https://doi.org/10.1186/1475-2875-10-144
Greenwood B (2014) Treatment of malaria—a continuing challenge. N Engl J Med 371(5):474–475. https://doi.org/10.1056/NEJMe1407026
Winzeler EA (2008) Malaria research in the post-genomic era. Nature 455(7214):751–756. https://doi.org/10.1038/nature07361
Gardner MJ et al (2002) Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419(6906):498–511. https://doi.org/10.1038/nature01097
Foth BJ et al (2008) Quantitative protein expression profiling reveals extensive post-transcriptional regulation and post-translational modifications in schizont-stage malaria parasites. Genome Biol 9(12):R177. https://doi.org/10.1186/gb-2008-9-12-r177
Raabe CA et al (2010) A global view of the nonprotein-coding transcriptome in Plasmodium falciparum. Nucleic Acids Res 38(2):608–617. https://doi.org/10.1093/nar/gkp895
Horrocks P et al (2009) Control of gene expression in Plasmodium falciparum—ten years on. Mol Biochem Parasitol 164(1):9–25. https://doi.org/10.1016/j.molbiopara.2008.11.010
Chene A et al (2012) PfAlbas constitute a new eukaryotic DNA/RNA-binding protein family in malaria parasites. Nucleic Acids Res 40(7):3066–3077. https://doi.org/10.1093/nar/gkr1215
Mani J et al (2011) Alba-domain proteins of Trypanosoma brucei are cytoplasmic RNA-binding proteins that interact with the translation machinery. PLoS ONE 6(7):e22463. https://doi.org/10.1371/journal.pone.0022463
Reddy BP et al (2015) A bioinformatic survey of RNA-binding proteins in Plasmodium. BMC Genomics 16:890. https://doi.org/10.1186/s12864-015-2092-1
Bell SD et al (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296(5565):148–151. https://doi.org/10.1126/science.1070506
Goyal M et al (2016) The Alba protein family: structure and function. Biochim Biophys Acta 1864(5):570–583. https://doi.org/10.1016/j.bbapap.2016.02.015
Tanaka T, Padavattan S, Kumarevel T (2012) Crystal structure of archaeal chromatin protein Alba2-double-stranded DNA complex from Aeropyrum pernix K1. J Biol Chem 287(13):10394–10402. https://doi.org/10.1074/jbc.M112.343210
Wardleworth BN et al (2002) Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J 21(17):4654–4662
Bell SD, Jackson SP (2001) Mechanism and regulation of transcription in archaea. Curr Opin Microbiol 4(2):208–213
Guo L et al (2014) Biochemical and structural insights into RNA binding by Ssh10b, a member of the highly conserved Sac10b protein family in Archaea. J Biol Chem 289(3):1478–1490. https://doi.org/10.1074/jbc.M113.521351
Goyal M et al (2012) Identification and molecular characterization of an Alba-family protein from human malaria parasite Plasmodium falciparum. Nucleic Acids Res 40(3):1174–1190. https://doi.org/10.1093/nar/gkr821
Duffy MF et al (2012) The role of chromatin in Plasmodium gene expression. Cell Microbiol 14(6):819–828. https://doi.org/10.1111/j.1462-5822.2012.01777.x
Bunnik EM, Le Roch KG (2015) PfAlba1: master regulator of translation in the malaria parasite. Genome Biol 16:221. https://doi.org/10.1186/s13059-015-0795-x
Vembar SS et al (2015) The PfAlba1 RNA-binding protein is an important regulator of translational timing in Plasmodium falciparum blood stages. Genome Biol 16:212. https://doi.org/10.1186/s13059-015-0771-5
Munoz EE et al (2017) ALBA4 modulates its stage-specific interactions and specific mRNA fates during Plasmodium yoelii growth and transmission. Mol Microbiol 106(2):266–284. https://doi.org/10.1111/mmi.13762
Zhu X-Q et al (2005) On-column refolding of an insoluble his6-tagged recombinant EC-SOD Overexpressed in Escherichia coli. Acta Biochim Biophys Sin 37(4):265–269. https://doi.org/10.1111/j.1745-7270.2005.00035.x
Tajiri M (2019) Comparison of high-throughput sequencing for phage display peptide screening on two commercially available platforms. Int J Pept Res Ther. https://doi.org/10.1007/s10989-019-09858-8
Blankenberg D et al (2010) Manipulation of FASTQ data with Galaxy. Bioinformatics 26(14):1783–1785. https://doi.org/10.1093/bioinformatics/btq281
Thiel WH, Giangrande PH (2016) Analyzing HT-SELEX data with the Galaxy Project tools—a web based bioinformatics platform for biomedical research. Methods 97:3–10. https://doi.org/10.1016/j.ymeth.2015.10.008
Stafford RL et al (2014) A general sequence processing and analysis program for protein engineering. J Chem Inf Model 54(10):3020–3032. https://doi.org/10.1021/ci500362s
Bailey TL, Machanick P (2012) Inferring direct DNA binding from ChIP-seq. Nucleic Acids Res 40(17):e128. https://doi.org/10.1093/nar/gks433
Dinkel H et al (2016) ELM 2016–data update and new functionality of the eukaryotic linear motif resource. Nucleic Acids Res 44(D1):D294-300. https://doi.org/10.1093/nar/gkv1291
Gouw M et al (2018) The eukaryotic linear motif resource—2018 update. Nucleic Acids Res 46(D1):D428–D434. https://doi.org/10.1093/nar/gkx1077
Bannister AJ, Kouzarides T (2011) Regulation of chromatin by histone modifications. Cell Res 21(3):381–395. https://doi.org/10.1038/cr.2011.22
Doerig C et al (2015) Post-translational protein modifications in malaria parasites. Nat Rev Microbiol 13(3):160–172. https://doi.org/10.1038/nrmicro3402
Hamilton MJ, Lee M, Le Roch KG (2014) The ubiquitin system: an essential component to unlocking the secrets of malaria parasite biology. Mol Biosyst 10(4):715–723. https://doi.org/10.1039/c3mb70506d
Ponts N et al (2011) Unraveling the ubiquitome of the human malaria parasite. J Biol Chem 286(46):40320–40330. https://doi.org/10.1074/jbc.M111.238790
Ye H et al (2002) Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418(6896):443–447. https://doi.org/10.1038/nature00888
Fu Y et al (2012) Plasmodium yoelii blood-stage primes macrophage-mediated innate immune response through modulation of toll-like receptor signalling. Malar J 11:104. https://doi.org/10.1186/1475-2875-11-104
Pozhidaeva A, Bezsonova I (2019) USP7: structure, substrate specificity, and inhibition. DNA Repair (Amst) 76:30–39. https://doi.org/10.1016/j.dnarep.2019.02.005
Deu E (2017) Proteases as antimalarial targets: strategies for genetic, chemical, and therapeutic validation. FEBS J 284(16):2604–2628. https://doi.org/10.1111/febs.14130
Ward P et al (2004) Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5:79. https://doi.org/10.1186/1471-2164-5-79
Dastidar EG et al (2012) Involvement of Plasmodium falciparum protein kinase CK2 in the chromatin assembly pathway. BMC Biol 10:5. https://doi.org/10.1186/1741-7007-10-5
Barro M, Patton JT (2005) Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc Natl Acad Sci USA 102(11):4114–4119. https://doi.org/10.1073/pnas.0408376102
Kanyal A et al (2017) Genome-wide survey and phylogenetic analysis of histone acetyltransferases and histone deacetylases of Plasmodium falciparum. FEBS J. https://doi.org/10.1111/febs.14376
Acknowledgements
I would like to thank Michigan Technological University Research Excellence Fund (R01547) and Portage Health Foundation (R75389). I would also like to thank Dr. Chelsea Nikula (National Physical Laboratory, London UK) for critical reviewing of the manuscript.
Funding
This work was funded by Michigan Technological University Research Excellence Fund (R01547) and Portage Health Foundation (R75389).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The author declares no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tajiri, M. Phage Display Screening for Alba Superfamily Proteins from the Human Malaria Parasite, Plasmodium falciparum Reveals a High Level of Association with Protein Modification Pathways and Hints at New Drug Targets. Acta Parasit. 66, 844–850 (2021). https://doi.org/10.1007/s11686-021-00339-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11686-021-00339-x